Journal of Oncobiomarkers
Download PDF
Review Article
*Address for Correspondence: Shunbin Ning, Viral Oncology Program, Division of Hematology/Oncology, Sylvester Comprehensive Cancer Center; Department of Cell Biology, Miller School of Medicine, University of Miami, Miami, Florida, 33136. E-mail: sning@med.miami.edu
Citation: Ning S. IRF4 as an Oncogenic Biomarker for Hematological Malignancies. J Oncobiomarkers. 2013;1(1): 6.
Copyright © 2013 Ning S. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Oncobiomarkers | ISSN: 2325-2340 | Volume: 1, Issue: 1
IRF4 as an Oncogenic Biomarker for Hematological Malignancies
Shunbin Ning*
- Viral Oncology Program, Division of Hematology/Oncology, Sylvester Comprehensive Cancer Center; Department of Cell Biology, Miller School of Medicine, University of Miami, Miami, Florida, 33136, USA
*Address for Correspondence: Shunbin Ning, Viral Oncology Program, Division of Hematology/Oncology, Sylvester Comprehensive Cancer Center; Department of Cell Biology, Miller School of Medicine, University of Miami, Miami, Florida, 33136. E-mail: sning@med.miami.edu
Citation: Ning S. IRF4 as an Oncogenic Biomarker for Hematological Malignancies. J Oncobiomarkers. 2013;1(1): 6.
Copyright © 2013 Ning S. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Oncobiomarkers | ISSN: 2325-2340 | Volume: 1, Issue: 1
Abstract
The lymphocyte-specific transcription factor Interferon (IFN) Regulatory Factor 4 (IRF4) is crucial for lymphocyte development. Importantly, IRF4 has potent oncogenic and transforming properties, and its intimate interaction with lymphoid and myeloid malignancies has been increasingly recognized. In general, IRF4 exerts its function by transcriptionally regulating a pool of genes pivotal for cell development, oncogenesis and immune response. In clinical practice, IRF4 serves as an important prognosis and diagnosis marker for certain types of these malignancies. However, the oncogenic roles of IRF4 in most types of these malignancies and the molecular mechanisms underlying its interaction with them are largely not characterized. Accumulating evidence from cell culture shows that IRF4 regulates differential targets in distinct cancer contexts, depending on the “context-specific” co-regulator(s) associated with it in each setting. These complementary studies with in vitro cell culture systems are a necessary strategy which will provide molecular and mechanistic insights into the specific regulation and function of IRF4 in distinct cancer contexts, and may identify novel interventions specifically targeting IRF4 regulatory network for treatment. This review summarizes the evidence obtained from bench to bed showing the association of IRF4 with various types of hematological malignancies, with emphasis on molecular mechanisms underlying its regulation and its roles in these contexts.Introduction
Interferon (IFN) Regulatory Factors (IRFs) are a small family of transcription factors which includes nine members in mammalians. IRFs play important roles in multiple facets of host defense systems especially in type I IFN production upon pathogenic infection [1], and are also involved in the regulation of tumorigene sis, cellgrowth, differentiation, and myeloid cell development [2]. All the IRF family members contain a conserved DNA binding domain (DBD) at their N-terminus, which binds to the consensus DNA sequences that generally comprises two GAAA repeats [3]. IRF4, also known as MUM1 (multiple myeloma (MM) oncoprotein 1), PIP (PU.1-interacting protein), LSIRF (Lymphocyte-specific IRF), ICSAT(Interferon consensus sequence binding protein for activated T cells), is a quintessential ‘context-dependent’ transcription factor whose DNA-binding specificity is profoundly shaped by lineagespecific transcriptional co-regulators, some of which have been identified, including PU.1, SPIB, DEF6, BATF, STAT3, NFAT and FKBP52 [4-13].IRF4 was cloned independently by different groups [4,14,15]. Its closest family member is IRF8, and together, they both play critical roles for the development of immune cells (lymphocytes, myelocytes, and dendritic cells). Irf4−/− mice are devoid of germinal centers (GCs) and plasma cells, with a severe reduction in the serum immunoglobulin (Ig) level and the failure of mounting protective antibody responses.
The expression of IRF4 is confined to immune cells including B cells, macrophages, and CD11b+ DCs, and is inducible by a variety of mitogenic stimuli including antigen receptor engagement, and TLR and CD40 signaling pathways [5]. IRF4 is strongly up regulated upon co-stimulation of B cells with CD40 and IL4. Commonly, these stimuli all activate NFκB leading to IRF4 induction [16]. Different from other IRF family members, expression of IRF4 is not induced by interferon. In B cell lineage, IRF4 expression is repressed by the transcription factor Mitf in mature B cells (native resting B cells), and is also particularly weak in GC B cells likely due to the absence of NFκB in these cells. The level of IRF4 protein is culminating in plasma cells, the terminally differentiated B cells, through a yet unknown mechanism. Among T cell lineages, its expression is the highest in activated T cells.
Like IRF2 [17] and -7 [18], IRF4 has oncogenic and transforming potentials and anti-apoptotic activity [19-21]. All these oncogenic IRFs intimately interact with Epstein-Barr Virus (EBV) latency programs [22], which are associated with a variety of hematological and epithelial malignancies.IRF4 is over expressed in a variety of hematological malignancies.
IRF4 over expression is a hallmark of ABC type of DLBCL and MM [5,23], and is frequently used as a diagnostic and prognostic marker for these and other proliferative disorders [5,23-26]. In these cancer contexts, IRF4 regulates cell cycle, apoptosis, and cell proliferation and survival [20,24-30], by transcriptionally targeting several key genes, including Blimp1, Ccnb1, Bcl6, Cdk6 and Myc [9,29,31,32]. These lines of evidence underscore the importance of IRF4 in these malignancies.
In this review, the association of IRF4 with multiple myeloma, DLBCL, and viral and other hematological malignancies is summarized and the known role and regulation of IRF4 in each cancer context are introduced.
IRF4 in multiple myeloma
Multiple myeloma(MM) is a malignancy of the terminally differentiated B lymphocytes, plasma cells, in which IRF4 is over expressed. Myc, which is over expressed owing to chromosomal translocation in this setting, induces expression of IRF4, and vice versa [29]. Chromosomal translocation also contributes to IRF4 over expression in a small fraction of MM [33]. Irf4 gene was identified as one of the six oncogenic chromosomal partners (the other five are Fgfr3, Ccnd3, Ccnd1, c-Maf, and Mafb) of IgH translocation in MM [33,34].
Transcriptome analysis has shown that IRF4 regulates a myeloma-specific gene expression pattern that combines IRF4 regulatory networks from activated B cells and plasma cells [29]. In addition to Myc, other important IRF4 targets in multiple myeloma include Casp3, Cdk6, Ell2, and Tnfaip3 [29]. Therefore IRF4 regulates multiple essential cellular processes such as cell cycle and apoptosis leading to myeloma cell proliferation and survival [27,29].
The Ets family members, PU.1 and its closely related factor SPIB, which are expressed in B cells, macrophages, and dendritic cells, are not expressed in MM [35]. Thus, IRF4 transcriptional co-regulator which recruits IRF4 to its genomic targets in MM remains to be determined.
IRF4 in DLBCL
DLBCL, a type of aggressive lymphoma with poor prognosis and the most common subtype of non-Hodgkin lymphoma, accounts for approximately 40% of lymphomas among adults. Over expression of IRF4 is a hallmark of ABC (activated B-cell-like) subtype of DLBCL [5,23], owing to the constitutive activation of the prosurvival NFκB pathway [32], a process mediated by the adapter CARD11 in DLBCL. The constitutive activation of NFκB results from oncogenic mutations of BCR and My D88. ABCDLBCL is likely originated from late germinal center B cells which fail to terminally differentiate into plasma cells. IRF4 over expression may manifest the physiological attempt of these cells to activate the terminal plasma cell differentiation program [36]. A recent report also shows that IRF4 is over expressed in a subtype of pediatric GCB-type DLBCL and follicular lymphoma grade 3 due to chromosomal translocation carrying Ig/Irf4 fusion loci [37].
In ABC DLBCL, IRF4 inhibits Irf7 gene transcription and therefore dampens type I IFN production, but stimulates NFκB signaling by transactivating Card11 gene promoter [35]. Like its role in MM, about 50% knockdown of IRF4 is sufficient to kill ABC DLBCL cell lines, but not to other tested lymphoma and leukemia lines [35]. Surprisingly, ChIP-Seq high throughput analysis has shown that, different from MM, IRF4 does not bind to Myc and Irf4 loci in ABC DLBCL lines, although MYC is highly expressed in ABC DLBCL, and that many other genes such as CD44 and CD40 are targeted by IRF4 in ABC DLBCL but not in MM [35]. These observations suggest distinct IRF4 regulatory networks in MM and DLBCL. In addition, unlike most normal GC B cells which display mutually exclusive in the expression of MUM1 and Bcl-6, tumor cells in approximately half of IRF4-positive DLBCL are also Bcl-6 positive. The IRF4 coregulator, SPIB that is not expressed in MM, intimately interacts with IRF4 for its oncogenic function in ABC DLBCL [35].
Nevertheless, different clinical studies have shown conflicts in terms of the prognostic value of IRF4 with DLBCL; some report poor prognostic value, but others have failed to observe any significance [38].
IRF4 in Burkitt lymphoma
Burkitt lymphoma (BL) is a type of non-Hodgkin B-cell lymphoma which is highly aggressive. BL is subdivided in to EBVassociated endemic subtype (>99% are EBV+), the sporadic subtype in developed countries (15-25% are EBV+), and an AIDS-associated subtype (~30% are EBV+) clinical variants [39]. BL is composed of monomorphic medium-sized B cells with a high proliferation rate, and c-Myc locus translocation as an oncogenic hallmark in all cases [40]. Classically, like DLBCL, BL is considered to be germinal center in immunophenotype [41]. In most reports, IRF4 is negative in BL cases. However, a study with 222 cases of well-characterized Burkitt lymphomas from Brazil with the classic phenotype and c-Myc translocation has revealed that 90 cases (40.5%) are IRF4-positive in the nucleus [41].
IRF4 and viral oncogenesis
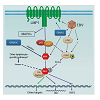
Virus contributes to appropriately 20% of total malignancies in the globe[42,43]. IRF4 is over expressed in EBV-transformed cells and associated lymphomas [20,44-46], and in Human T-cell Leukemia Virus-1 (HTLV1)-transformed cell lines and associated Adult T-cell Lymphoma/Leukemia (ATLL) [31,47-50]. It is now understood that, in EBV-transformed cells, IRF4 is induced by the LMP1/NFκB signaling (Figure 1) [20,46]. Consistent with this, IRF4 has been recently shown to be expressed in all EBV LMP1-driven tumors in mice [51]. More recently, IRF4 has been shown to be stabilized by EBNA3 Cin EBV-transformed cells (Figure 1) [52]. An over expression study also showed that IRF4 is a direct target for the EBV antigen EBNA2, as detected by microarray analysis (Figure 1) [53]. Induction of IRF4 expression in the context of HTLV1 infection is complicated. It is induced in a Tax-independent manner in primary ATLL [54], but is induced in either Tax-dependent or Taxindependent manner in cell culture [47,49,54]. Induction of IRF4 expression by Tax may be an important cellular event involved in HTLV1 leukemogenesis [50]. IRF4 can also be induced by c-Rel or by other undefined cellular pathways in the absence of c-Rel or Tax [47,54].
LMP1 is the principal EBV oncogene, and is the only EBV product which transforms human and rodent fibroblasts in vitro [55,56]. Up to 50% of LMP1 transgenic (Tg) mice had a threefold increase in lymphoma development in elderly period in comparison with LMP1-negative mice [57]. Moreover, on a T-cell ablated background, most LMP1 Tg mice developed DLBCL-like and plasmacytic tumors [51], As such, IRF4 can induce focus formation of mouse fibroblasts in soft agar, a typical sign of malignant transformation [20]. The importance of IRF4 in LMP1 oncogenesis is underscored by the fact that IRF4 is expressed in all of these LMP1-driven tumors in mice [51].
Nevertheless, the role of IRF4 in EBV/LMP1 oncogenesis is unclear. Importantly, we have recently identified B-cell integration cluster (BIC) as the first miRNA-encoding gene induced by IRF4 in virus-transformed cells (Figure 1) [21]. BIC encodes miR-155, which plays important roles in innate immunity [58,59], and is the first identified oncogenic miRNA (oncomiR) implicated in various types of cancers including lymphomas [60-62], breast cancer, leukemia, pancreatic cancer, and lung cancer [63,64]. Targeted expression of miR-155 alone in B cells developed B cell malignancies in transgenic mice [65], and enforced expression in mouse bone marrow cells causes myeloid neoplasia [66]. As an important miRNA in immunity and cancer, miR-155 preferentially targets SHIP1 [67], among many others [68-70]. Like oncogenic IRFs, miR-155 is also associated with EBV latency [61,71-73. BIC/miR-155 is induced by TLR signaling, TNFα, IFN-β, IFN-γ, EBV LMP1, LMP2A [74] and B cell receptor (BCR) engagement. However, little is known about the mechanism controlling its regulation [75]. Therefore, our findings unveiled an intersection between two major and quite diverse models in the regulation of viral oncogenesis, and have provided valuable insights into the interaction between viral oncogenesis and immune mechanisms governed by them. For example, both factors are crucial regulators of germinal center reaction [36,76], which is implicated in lymphoma development and EBV latent infection [77]. Also, IRF4 has been shown to repress IRF5 expression in EBV-transformed cells (Figure 1) [78].
Furthermore, our microarray analysis shows that IRF4 regulates a pool of interesting genes in the context of EBV infection, including a subgroup of the genes such as Cdk6 and Ccnb1 which are also targeted by IRF4 in MM (data not published). Future pursuits on selected targets may unravel novel and specific roles for IRF4 in EBV oncogenesis, and broaden our knowledge in its interaction with viral oncogenesis and other associated cancers.
Figure 1: A tentative scheme for IRF4 regulation in EBV latency. IRF4 expression is induced by the LMP1/NFκB axis [20;44-46], and probably is also directly induced by EBNA2 [53]. IRF4 protein is also stabilized by EBNA3C [52]. Our recent study has disclosed that IRF4 is activated through c-Srcmediated tyrosine phosphorylation [91]. We also have evidence showing that IRF4 is activated by the LMP1 signaling pathway, probably mediated by c-Src. c-Src protein is induced and activated in EBV latency [91]. In addition to the LMP1 pathway, other lymphocyte-specific pathways may also contribute to IRF4 activation. Two transcriptional targets, BIC [21] and Irf5 [78], have been identified and confirmed so far for IRF4 in EBV latency. IRF4 induces BIC but represses Irf5 transcription.
IRF4 and other hematological malignancies
In addition to MM, chromosomal translocation and genetic mutation of IRF4 have been found in peripheral T-cell lymphomas [79], and Chronic Lymphocytic Leukemia (CLL) [33,80]. Also, IRF4 over-expression was found in various types of T cell lymphomas in addition to ATLL, in most Hodgkin lymphoma cases, and in follicular lymphoma, primary effusion lymphoma(PEL), primary central nervous system lymphoma, and anaplastic large cell lymphoma (ALCL) [16,53,81], as well as in the context of the acquired immunodeficiency syndrome (AIDS) and in post-transplant lymphoproliferative disorders (PTLD) [41]. IRF4 may serve as one of the phenotypic markers of B-cell lymphoma histogenesis. In particular, IRF4 may be a marker for the transition from Bcl6+ germinal center B cell (GCB) to CD138+ immunoblasts and plasma cells [16]. IRF4 may help in the discrimination of PEL versus other lymphomas involving the serous body cavities, which are usually IRF4 negative [16].
In addition to hematological malignancies, an interesting report has shown that the germline variant IRF4 rs12203592 T allele was associated with increased risks of several types of skin cancers including melanoma, squamous cell carcinoma and basal cell carcinoma [82].
IRF4 as a tumor suppressor
IRF4, when over-expressed, plays an important role in the pathogenesis of hematopoietic malignancies. However, in some certain cancer contexts such as B-cell acute lymphoblastic leukemia (B-ALL), chronic myeloid leukemia (CML), acute myeloid leukemia (AML), and chronic myelomonocytic leukemia, IRF4 is down regulated. In these cases, IRF4 acts as a tumor suppressor [83-85]. In Irf4+/- heterozygous mice, c-Myc-induced leukemia was significantly accelerated [84]. It is also notable that IRF4 plays a dual role for lymphocyte activation/development and death by summating distinct incoming signals [30,86], and thus plays a central role in integrating the life and death decisions for lymphocytes [87].
Activation of IRF4 in cancer
As a transcription factor, activation of IRF4 is prerequisite for its function. Serine phosphorylation of IRF4 by the kinase ROCK2 activates IRF4 leading to IL17/21 production in autoimmune response in mice [88]. However, how IRF4 is activated in cancer is an open question which has never been mentioned. Many proteins involved in cancer signal transduction are tyrosine-phosphorylated. A few limited high throughput profiling studies have identified several tyrosine phosphorylation sites on IRF4 in different cancer contexts, including Y192 in MM [89], and Y37, Y122, Y125, Y428 and Y440 in Hodgkin lymphomas [90]. Interestingly, our recent phospho-proteome analysis has shown that IRF4 is also tyrosine phosphorylated in EBV-transformed cells, and identified several phosphorylation sites including Y125, Y192, Y181 and Y428 on IRF4. We have concluded that Y125 is a promising phosphorylation site at least important for IRF4-associated lymphomas [88]. We have further shown that the tyrosine kinase c-Src promotes IRF4 phosphorylation and activation, and identified Y62 and Y125 of IRF4 as two key sites responding to c-Src-mediated activation (Figure 1) [91]. Moreover, we show that c-Src is constitutively expressed and activated in EBV transformed cells [91]. These findings indicate that IRF4 is activated through a c-Src-mediated pathway in EBV-transformed cells. However, our data suggests that c-Src is unlikely a direct kinase for IRF4 [91]. Further study will be followed to confirm this claim and to identify the direct kinase(s).
Discussion and Prospectives
Increasing evidence has shown the implication of IRF4 deregulation in diverse hematological malignancies. IRF4 serves as aprognosis marker for MM and ABC DLBCL and was also proposed as a prognosis marker for classical Hodgkin lymphoma [92]. IRF4 overexpression has been found in almost 100% cases of MM, classical Hodgkin lymphoma, primary central nervous system DLBCL, plasma cell myeloma and PEL. In many cases, IRF4 may be used as an important marker in combination with other markers for diagnosis. For example, for follicular lymphoma and DLBCL, IRF4 can be used with CD10 and Bcl6 for diagnosis [16].However, the functional roles of IRF4 and the mechanisms underlying its interaction with these cancers remain to be elucidated. Study with cell culture systems is a necessary tool for this purpose. Targeted expression of IRF4 in mouse lymphocytes failed to develop any cancer [93], probably due to the fact that IRF4 requires co-factors for its function. Thus, gene-targeted mouse models are necessary to be established for the in vivo study of the interaction between IRF4 and other factors such as LMP1 in developing malignancies.
Currently, important work includes systematic and in-depth analyses of functional roles of IRF4 in distinct cancer contexts, the identification of “context-dependent” co-factors for IRF4, and the identification of potential lymphocyte-specific signaling pathways leading to IRF4 activation in each cancer context (Figure 1).These studies will highlight the importance of IRF4 in the pathogenesis of these cancers and will establish IRF4 as a unique therapeutic target for treating these hematological malignancies and other cancers [5]. Since IRF4 is a lineage-dependent transcription factor, discriminating its specific roles in different cellular developmental stages and different cancer contexts may provide unique opportunities to target IRF4 regulatory network for treating these diverse proliferative diseases. Finally, we envision discovering novel signaling pathways and novel molecules such as the kinase(s) for IRF4 which may open up unique opportunities for therapeutic treatments of IRF4-associated hematological malignancies.
Acknowledgements
This work is supported by the State of Florida Biomedical Research Programs (1BN-07) and American Society of Hematology Scholar Award to S. N.References
- Taniguchi T, Takaoka A (2002) The interferon-a/b system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr Opin Immunol 14:111-116.
- Yanai H, Negishi H, Taniguchi T (2012) The IRF family of transcription factors: Inception, impact and implications in oncogenesis. Oncoimmunology 1:1376-1386.
- Chen W, Royer W (2010) Structural insights into interferon regulatory factor activation. Cell.Signal 22: 883-887.
- Eisenbeis CF, Singh H, Storb U (1995) Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9: 1377-1387.
- Shaffer AL, Emre NC, Romesser PB, Staudt LM. 2009. IRF4: immunity. malignancy! therapy? Clin Cancer Res 15: 2954-2961.
- Mamane Y, Sharma S, Petropoulos L, Lin R, Hiscott J (2000) Posttranslational regulation of IRF-4 activity by the immunophilin FKBP52. Immunity 12: 129-140.
- Biswas PS, Bhagat G, Pernis AB (2010) IRF4 and its regulators: evolving insights into the pathogenesis of inflammatory arthritis? Immunol Rev 233: 79-96.
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, et al (2012) A genomic regulatory element that directs assembly and function of immunespecific AP-1-IRF complexes. Science 16: 975-980.
- Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, et al. (2009) Analysis of Interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31: 941-952.
- Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, KC W, et al. (2012) Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490: 502-507.
- Li P, Spolski R, Liao W, Wang L, Murphy TL, et al. (2012) BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 490: 543-546.
- Farrow MA, Kim EY, Wolinsky SM, Sheehy AM (2011) NFAT and IRF proteins regulate transcription of the anti-HIV protein, APOBEC3G. J.Biol Chem 286: 2567-2577.
- Murphy TL, Tussiwand R, Murphy KM (2013) Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol 13: 499-509.
- Matsuyama T, Grossman A, Mittrucker HW, Siderovski DP, Kiefer F, et al. (1995) Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE). Nucleic Acids Res. 23: 2127-2136.
- Yamagata T, Nishida J, Tanaka T, Sakai R, Mitani K, et al. (1996) A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol Cell Biol 16: 1283-1294.
- HeGualco G, Weiss LM, Bacchi CE (2010) MUM1/IRF4: A review Appl. Immunohistochem. Mol Morphol 18: 301-310.
- Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, et al. (1993) Anti-oncogenic and oncogenic potentials of interferon regulatory factor-1 and factor-2. Science 259: 971-974.
- Zhang L, Zhang J, Lambert Q, Der CJ, Del Valle L, et al. (2004) Interferon regulatory factor 7 is associated with Epstein-Barr virus-transformed central nervous system lymphoma and has oncogenic properties. J Virol 78: 12987-12995.
- Lohoff M, Mittrucker HW, Brustle A, Sommer F, Casper B, et al. (2004) Enhanced TCR-induced apoptosis in interferon regulatory factor 4-deficient CD4(+) Th cells. J Exp Med 200: 247-253.
- Xu D, Zhao L, Del Valle L, Miklossy J, Zhang L (2008) Interferon regulatory factors 4 is involved in Epstein-Barr virus-mediated transformation of human B lymphocytes. J Virol 82: 6251-6258.
- Wang L, Toomey NL, Diaz LA, Walker G, Ramos JC, et al. (2011) Oncogenic IRFs provide a survival advantage for EBV- or HTLV1-transformed cells through induction of BIC expression. J Virol 85: 8328-8337.
- Ning S, Pagano J, Barber G (2011) IRF7: activation, regulation, modification, and function. Genes & Immun 12: 399-414.
- Rui L, Schmitz R, Ceribelli M, Staudt LM (2011) Malignant pirates of the immune system. Nat.Immunol 12: 933-940.
- Sundram U, Harvell JD, Rouse RV, Natkunam Y (2003) Expression of the B-cell proliferation marker MUM1 by melanocytic lesions and comparison with S100, gp100 (HMB45), and MelanA. Mod Pathol 16: 802-810.
- Chang CC, Lorek J, Sabath DE, Li Y, Chitambar CR, et al. (2002) Expression of MUM1/IRF4 correlates with clinical outcome in patients with B-cell chronic lymphocytic leukemia. Blood 100: 4671-4675.
- Heintel D, Zojer N, Schreder M, Strasser-Weippl K, Kainz B, et al. (2007) Expression of MUM1//IRF4 mRNA as a prognostic marker in patients with multiple myeloma. Leukemia 22: 441-445.
- Huang X, Di Liberto M, Jayabalan D, Liang J, Ely S, et al. (2012) Prolonged early G1 arrest by selective CDK4/CDK6 inhibition sensitizes myeloma cells to cytotoxic killing through cell cycle-coupled loss of IRF4. Blood 120: 1095-1106.
- Aldinucci D, Celegato M, Borghese C, Colombatti A, Carbone A (2011) IRF4 silencing inhibits Hodgkin lymphoma cell proliferation, survival and CCL5 secretion. British Journal of Haematology 152: 182-190.
- Shaffer AL, Emre NCT, Lamy L, Ngo VN, Wright G, et al. (2008) IRF4 addiction in multiple myeloma. Nature 454: 226-231.
- Fanzo JC, Hu CM, Jang SY, Pernis AB (2003) Regulation of lymphocyte apoptosis by interferon regulatory factor 4 (IRF-4). J Exp Med 197: 303-314.
- Mamane Y, Grandvaux N, Hernandez E, Sharma S, Innocente SA, etal. (2002) Repression of IRF-4 target genes in human T cell leukemia virus-1 infection. Oncogene 21: 6751-6765.
- Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, etal. 2007. A signaling pathway mediating down regulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell 12: 280-292.
- Iida S, Rao PH, Butler M, Corradini P, Boccadoro M, et al (1997) Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet 17: 226-230.
- Yoshida S, Nakazawa N, Iida S, Hayami Y, Sato S, et al. (1999) Detection of MUM1/IRF4-IgH fusion in multiple myeloma. Leukemia 13: 1812-1816.
- Yang Y, Shaffer III AL, Emre NC, Ceribelli M, Zhang M, etal. 2012. Exploiting synthetic lethality for the therapy of ABC Diffuse Large B Cell Lymphoma. Cancer Cell 21: 723-737.
- De Silva NS, Simonetti G, Heise N, Klein U (2012) The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol.Rev 247: 73-92.
- Salaverria I, Philipp C, Oschlies I, Kohler CW, Kreuz M, et al. (2011) Translocations activating IRF4 identify a subtype of germinal center-derived B-cell lymphoma affecting predominantly children and young adults. Blood 118: 139-147.
- Bodoor K, Matalka I, Hayajneh R, Haddad Y, Gharaibeh W (2012) Evaluation of BCL-6, CD10, CD138 and MUM-1 expression in diffuse large B-cell lymphoma patients: CD138 is a marker of poor prognosis. Asian Pac J Cancer Prev. 13: 3037-3046.
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, etal. 2012. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490: 116-120.
- Allday MJ (2009) How does Epstein-Barr virus (EBV) complement the activation of Myc in the pathogenesis of Burkitt’s lymphoma? Semin Cancer Biol 19: 366-376.
- Gualco G, Queiroga EM, Weiss LM, Klumb CE, Harrington WJ, et al. (2009) Frequent expression of multiple myeloma 1/interferon regulatory factor 4 in Burkitt lymphoma. Hum Pathol 40: 565-571.
- Blossom D (2007) DNA tumor viruses and human cancer. Trends in Microbiology 15: 38-44.
- Goedert JJ (2000) Infectious causes of cancer: targets for intervention. Humana Press.
- Martin HJ, Lee JM, Walls D, Hayward SD (2007) Manipulation of the Toll-Like Receptor 7 signaling pathway by Epstein-Barr Virus. J Virol 81: 9748-9758.
- Spender LC, Lucchesi W, Bodelon G, Bilancio A, Karstegl CE, et al. (2006) Cell target genes of Epstein-Barr virus transcription factor EBNA-2: induction of the p55a regulatory subunit of PI3-kinase and its role in survival of EREB2.5 cells. J GenVirol 87: 2859-2867.
- Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, et al. (2004) Role of NF-kB in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr Virus latency III-infected cells. J Virol 78: 4108-4119.
- Ramos JC, Ruiz P, Ratner L, Reis IM, Brites C, et al. (2007) IRF4 and c-Rel expression in antiviral-resistant adult T-cell leukemia/lymphoma. Blood 109: 3060-3068.
- Sharma S, Grandvaux N, Mamane Y, Genin P, Azimi N, et al. (2002) Regulation of IFN regulatory factor 4 expression in human T cell leukemia virus-I-transformed T cells. J Immunol 169: 3120-3130.
- Sharma S, Mamane Y, Grandvaux N, Bartlett J, Petropoulos L, et al. (2000) Activation and regulation of interferon regulatory factor 4 in HTLV type 1-infected T lymphocytes. AIDS Res Hum Retrov 16: 1613-1622.
- Mamane Y, Sharma S, Grandvaux N, Hernandez E, Hiscott J (2002) IRF-4 activities in HTLV-I-induced T cell leukemogenesis. J Interferon Cytokine Res. 22: 135-143.
- Zhang B, Kracker S, Yasuda T, Casola S, Vanneman M, et al. (2012) Immune surveillance and therapy of lymphomas driven by Epstein-Barr Virus protein LMP1 in a mouse model. Cell 148: 739-751.
- Banerjee S, Lu J, Cai Q, Saha A, Jha HC, et al. (2013) The EBV latent antigen 3C inhibits apoptosis through targeted regulation of Interferon Regulatory Factors 4 and 8. PLoS Pathog 9: e1003314.
- Arguello M, Sgarbanti M, Hernandez E, Mamane Y, Sharma S, et al. (2003) Disruption of the B-cell specific transcriptional program in HHV-8 associated primary effusion lymphoma cell lines. Oncogene 22: 964-973.
- Suzuki S, Zhou Y, Refaat A, Takasaki I, Koizumi K, et al. (2010) Human T Cell Lymphotropic Virus 1 manipulates interferon regulatory signals by controlling the TAK1-IRF3 and IRF4 pathways. J Biol Chem 285: 4441-4446.
- Mainou BA, Everly DN, Raab-Traub N (2005) Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917-6924.
- Mainou BA, Raab-Traub N (2006) LMP1 strain variants: biological and molecular properties. J Virol 80: 6458-6468.
- Kulwichit W, Edwards RH, Davenport EM, Baskar JF, Godfrey V, et al. (1998) Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc Natl Acad.Sci USA 95: 11963-11968.
- Pedersen I, David M (2008) MicroRNAs in the immune response. Cytokine 43:391-394.
- Xiao C, Rajewsky K (2009) MicroRNA control in the immune system: basic principles. Cell 136: 26-36.
- van den BA, Kroesen BJ, Kooistra K, de JD, Briggs J, et al. (2003) High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes.Cancer 37:20-28.
- Kluiver J, Haralambieva E, de JD, Blokzijl T, Jacobs S, et al. (2006) Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer 45: 147-153.
- Lin Z, Flemington EK (2011) miRNAs in the pathogenesis of oncogenic human viruses. Cancer Lett 305: 186-199.
- Garzon R, Croce CM (2008) MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol 15: 352-358.
- Cho WC (2007) OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer 6: 60.
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, et al. (2006) Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc.Natl.Acad.Sci U.S.A 103:7024-7029.
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, et al. (2008) Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med 205: 585-594.
- O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. 2009. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA 106: 7113-7118.
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, et al. (2007) Requirement of bic/microRNA-155 for normal immune function. Science 316: 608-611.
- Tili E, Croce CM, Michaille JJ (2009) miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol 28: 264-284.
- Faraoni I, Antonetti FR, Cardone J, Bonmassar E (2009) miR-155 gene: A typical multifunctional microRNA. Biochimica et Biophysica Acta 1792: 497-505.
- Cameron JE, Fewell C, Yin Q, McBride J, Wang X, et al. (2008) Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology 382: 257-266.
- Jiang J, Lee EJ, Schmittgen TD (2006) Increased expression of microRNA-155 in Epstein-Barr virus transformed lymphoblastoid cell lines. Genes Chromosomes.Cancer 45:103-106.
- Yin Q, McBride J, Fewell C, Lacey M, Wang X, et al. (2008) MicroRNA-155 is an Epstein-Barr Virus-induced gene that modulates Epstein-Barr Virusregulated gene expression pathways. J Virol 82: 5295-5306.
- Du ZM, Hu LF, Wang HY, Yan LX, Zeng YX, et al. (2011) Upregulation of miR-155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS One 6:e19137.
- Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, et al. (2008) Epstein-Barr virus latent membrane protein 1 transactivates miR-155 transcription through the NF-kB pathway. Nucleic Acids Res. 36: 6608-6619..
- Thai T-H, Calado DP, Casola S, Ansel KM, Xiao C, et al. (2007) Regulation of the germinal center response by microRNA-155. Science 316:604-608.
- Roughan JE, Thorley-Lawson DA (2009) The intersection of Epstein-Barr Virus with the germinal center. J Virol 83: 3968-3976.
- Xu D, Meyer F, Ehlers E, Blasnitz L, Zhang L (2011) Interferon regulatory factor 4 (IRF-4) targets IRF-5 to regulate Epstein-Barr virus transformation. J Biol Chem 286: 18261-18267.
- Feldman AL, Law M, Remstein ED, Macon WR, Erickson LA, et al. (2009)Recurrent translocations involving the IRF4 oncogene locus in peripheral T-cell lymphomas. Leukemia 23:574-580.
- Havelange V, Pekarsky Y, Nakamura T, Palamarchuk A, Alder H, et al. (2011) IRF4 mutations in chronic lymphocytic leukemia. Blood 118: 2827-2829.
- Tsuboi K, Iida S, Inagaki H, Kato M, Hayami Y, et al. (2000) MUM1/IRF4 expression as a frequent event in mature lymphoid malignancies. Leukemia 14: 449.
- Han J, Qureshi AA, Nan H, Zhang J, Song Y, et al. (2011) A germline variant in the Interferon Regulatory Factor 4 gene as a novel skin cancer risk locus. Cancer Res 71: 1533-1539.
- Acquaviva J, Chen X, Ren R (2008) IRF4 functions as a tumor suppressor in early B-cell development. Blood 112: 3798-3806.
- Pathak S, Ma S, Trinh L, Eudy J, Wagner KU, et al. (2011) IRF4 is a suppressor of c-Myc induced B cell leukemia. PLoS.ONE. 6: e22628.
- Teng Y, Takahashi Y, Yamada M, Kurosu T, Koyama T, et al. (2007) IRF4 negatively regulates proliferation of germinal center B cell-derived Burkitt’s lymphoma cell lines and induces differentiation toward plasma cells. Eur J Cell Biol 86: 581-589.
- Mittrucker HW, Matsuyama T, Grossman A, Kundig TM, Potter J, et al. (1997) Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science 275:540-543.
- Pernis AB (2002) The role of IRF-4 in B and T cell activation and differentiation. J.Interferon Cytokine Res. 22: 111-120.
- Biswas PS, Gupta S, Chang E, Song L, Stirzaker RA, et al. (2010) Phosphorylation of IRF4 by ROCK2 regulates IL-17 and IL-21 production and the development of autoimmunity in mice. J Clin Invest 120: 3280-3295.
- St-Germain JR, Taylor P, Tong J, Jin LL, Nikolic A, et al. (2009) Multiple myeloma phosphotyrosine proteomic profile associated with FGFR3 expression, ligand activation, and drug inhibition. Proc Natl Acad Sci USA 106: 20127-20132.
- Gu TL, Cherry J, Tucker M, Wu J, Reeves C, et al. (2010) Identification of activated Tnk1 kinase in Hodgkin’s lymphoma. Leukemia 24: 861-865.
- Wang L, Ning S (2013) IRF4 is activated through c-Src-mediated tyrosine phosphorylation in virus-transformed cells. J Virol 87: In press.
- Valsami S, Pappa V, Rontogianni D, Kontsioti F, Papageorgiou E, et al. (2007) A clinicopathological study of B-cell differentiation markers and transcription factors in classical Hodgkin’s lymphoma: a potential prognostic role of MUM1/IRF4. Haematologica 92:1343-1350.
- Saito T, Yamagata T, Takahashi T, Honda H, Hirai H (1999) ICSAT overexpression is not sufficient to cause adult T-cell leukemia or multiple myeloma. Biochem Biophys Res Commun 260: 329-331.