Advances in Diabetes & Endocrinology
Download PDF
Case Report
Genetic Polymorphism and Risk of having Type 2 Diabetes in a Palestinian Population: A Study of 16 Gene Polymorphisms
Fadel A. Sharif*, Mohammed E. Shubair, Mazen M. Zaharna, Mohammed J. Ashour, Israa O. Altalalgah, Mahmoud Najjar and Mohammed Thalathini
- Department of Medical Laboratory Sciences, Islamic University ofGaza, Palestine
*Address for Correspondence: Fadel A. Sharif, Department of Medical Laboratory Sciences, Islamic Universityof Gaza, Gaza, Palestine.; E-mail: fsharif@iugaza.edu.ps; fadelsharif@gmail.com
Citation: Sharif FA, Shubair ME, Zaharna MM, Ashour MJ. Altalalgah IO, et al. Genetic Polymorphism and Risk of having Type 2 Diabetes in a Palestinian Population: A Study of 16 Gene Polymorphisms. Adv Diabetes Endocrinol 2018;3(1): 6.
Copyright: © 2018 Sharif FA, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Advances in Diabetes & Endocrinology | ISSN: 2475-5591 | Volume: 3, Issue: 1
Submission: January 10, 2018| Accepted: February 05, 2018 | Published: : February 12, 2018
Abstract
Background: Type 2 Diabetes (T2D) is a multifactorial disease that encompasses environmental risk factors and the contribution of multiple genomic variants. Studies on the genetic components of T2D revealed many T2D-associated genetic polymorphisms in various populations. Lack of studies on the relation between gene polymorphism and T2D in Palestinians prompted us to examine the association between 16 known single nucleotide polymorphisms (SNPs) and T2D in this unexplored population.
Method: In this case-control study, 100 T2D male patients and 100 control men were examined. The two groups were genotyped for the 16 genes polymorphisms using PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) technique. Body Mass Index (BMI), and essential clinical parameters were measured for all the study participants. The relation between the 16 SNPs and T2D were statistically analyzed using appropriate tests.
Results: Significant association was evident between IGF2BP and T2D, followed by CDKN2A/B (rs10811661) (OR=2.35, P-value=0.003), COL8A1 (rs792837) (OR=2.03, P-value=0.015), KCNQ1 (rs2237892) (OR=0.184, P-value=0.017), and KCNJ11 (rs5219; E23K) (OR=1.81, P-value=0.04), based on Armitage trend test. Among the 16 tested polymorphisms, a highly statistically significant association was evident between IGF2BP2 (rs4402960) and T2D [Odds ratio (OR)=3.28, P-value=7.46x10-8].
Conclusion: IGF2BP, CDKN2A/B, COL8A1, KCNQ1, and KCNJ11 gene variants are associated with T2D in the investigated population. This preliminary study sheds some light on the genetic components of T2Din Palestine.
Keywords
Type-2 Diabetes; Gene; Single Nucleotide Polymorphism; Palestine
Introduction
Type 2 Diabetes (T2D) is a global health concern with more than 300 million patients worldwide, and its prevalence is continuing to escalate in many populations [1], including Palestine. The estimated prevalence of diabetes in Palestinians, by the year 2010, was around 14% [2].
T2D is multifactorial disease that develops and progresses as a result of interaction between multiple genomic (both genetic and epigenetic) variants and various environmental factors [3,4].
Genetic alterations, in the form of single nucleotide polymorphisms (SNPs), in more than 80 loci have been associated with the susceptibility of having T2D in various populations, Caucasians in particular [5,6]. Functional studies have shown that many of those loci are related to the main aspects of T2D pathophysiology namely, insulin secretion and insulin resistance (and its underlying obesity) in peripheral tissues [7].
The gene polymorphisms investigated in the present study and their potential pathomechanisms in T2D were previously reported [8,12]. Earlier studies have shown that, at least for certain genetic polymorphisms, gender influences the outcome of association with T2D and that men and women genotypes should be assessed separately [13,14].
The Gaza strip is a small area (365 km2) located in the southwestern part of Palestine. It lies at the Mediterranean southeast coast. The strip is inhabited by around 2 million people.
Palestinians genetic susceptibility to T2D is largely unexplored, therefore this preliminary study was undertaken in order to identify some of the genetic risk alleles at reported SNPs.
Methods
Ethics approval and consent to participate
Written informed consent was obtained from all participants, and approval for conducting the study was obtained from the localHelsinki ethics committee.
Subjects
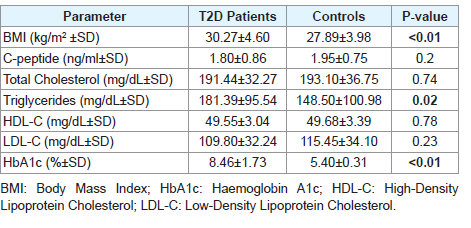
Two hundred males, including 100 unrelated T2D patients and 100 controls without diabetes, were enrolled in this study. T2D patients were recruited from Governmental Diabetes clinics in Gaza strip. BMI and relevant clinical parameters of the study participants are indicated in [Table 1]. All subjects were in the age range of 35 to 50 years.
DNA extraction
The DNA was isolated from whole blood samples using Wizard DNA extraction kit (Promega, USA) as described by the manufacturer.
SNP Selection and Genotyping
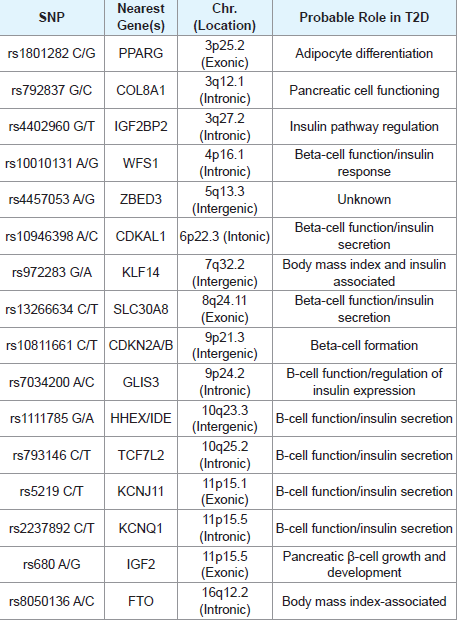
Sixteen SNPs validated for association with T2D in other populations have been investigated in this study [Table 3]. SNPs were selected on basis of their consistent association with T2D reported in diverse populations [8,12].
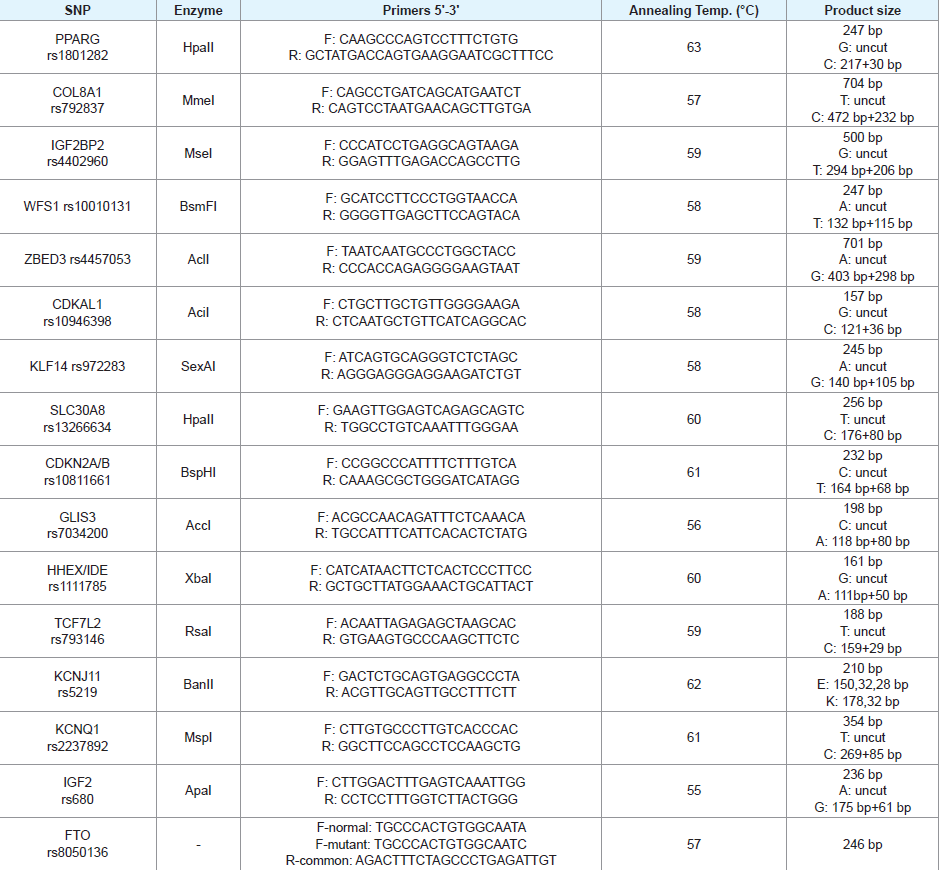
PCR-RFLP was used for genotyping the selected SNPs. The PCR primers for genotyping COL8A1, WFS1, ZBED3, KLF14, and GLIS3 SNPs were designed using online Primer3 software (http:// primer3.ut.ee/) based on the genomic sequence of each SNP (http:// www.ncbi.nlm.nih.gov/snp/). Then restriction enzymes required for the PCR-RFLP identification of each SNP were selected from New England Biolabs (NEB) database by using NEBcutter V2.0 online software (http://nc2.neb.com/NEBcutter2/). Genotyping of the FTO polymorphism was carried out using allele-specific PCR. Primers, enzymes, and reaction conditions for the rest of the SNPs were as previously reported [15-23,32]. Sequences of the PCR primers employed in this work are presented in [Table 3]
PCR was performed to amplify each required fragments. In a reaction mixture of 20 µL containing: 10 µL master mix (Promega,USA), 10 mmol/L of each primer, and 100ng genomic DNA. A typical 35 amplification cycles consisted of 30 seconds at 96 °C, 45 seconds at annealing temperature [Table 3], and 30 seconds at 72 °C. The final elongation step was 5 minutes at 72 °C. Restriction enzyme digestion was carried out as instructed by the manufacturer (NEB, Ipswich, MA, USA). The restricted PCR products were visualized by electrophoresis in 3% agarose gel stained with ethidium bromide. Alleles and genotypes were assigned according to the product sizes indicated in [Table 3].
Statistical analysis
The genotype/allele frequencies in T2D patients and the controls were analyzed by standard Chi-square test. Logistic regression was used for computing “unadjusted” odds ratios and their corresponding 95% confidence intervals (CIs) considering diabetes as the dependent variable and the genotypes as independent variables. Student’s t-test was used to evaluate the differences of the continuous variables (presented as mean ± standard deviation) between cases and controls. Hardy-Weinberg equilibrium (HWE) was tested using freely available software (http://www.oege.org/software/hwe-mr-calc. shtml). The Armitage trend test was applied to assess for the presence of association between the different groups of genotypes.
Results
Genotype and allele frequencies of investigated polymorphisms
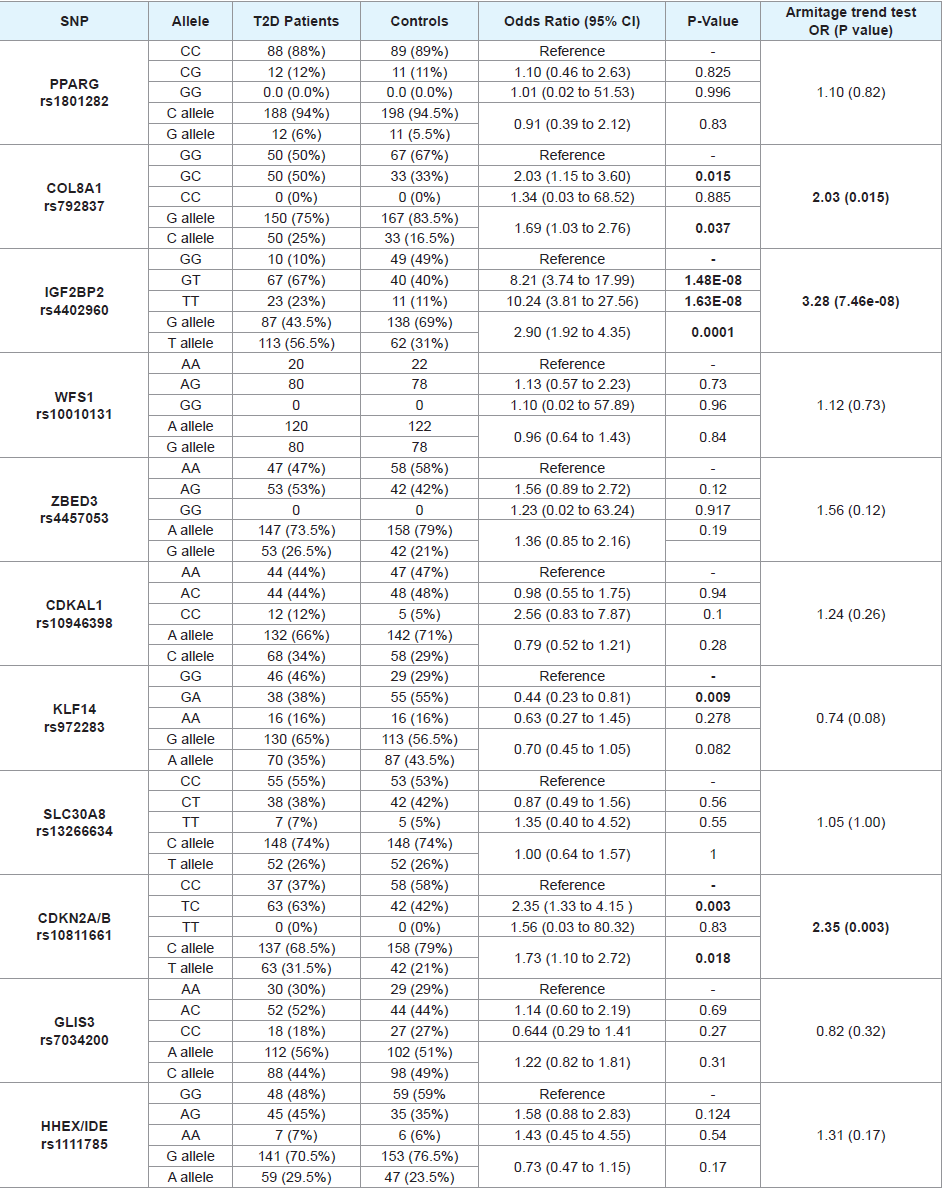
[Table 4] illustrates genotype/allele frequencies, odds ratios, 95% confidence intervals, crude P values and Armitage P trend values for the tested genes’ polymorphisms among T2D patients and controls. Statistical analyses of genotypic and allelic frequencies for the tested SNPs revealed significant trend (all P trend values are <0.05) difference between T2D patients and controls in 5 of the tested genes (COL8A1, IGF2BP2, CDKNA2A/B, KCNJ11, and KCNQ1) polymorphisms. The remaining SNPs do not seem to impact T2D risk in the investigated population.
Apart from WFS1 (rs10010131), ZBED3 (rs4457053) and CDKNA2A/B (rs10811661) genotypes, which showed the corresponding P-values for those three SNPs were: <0.001, 0.008, and 0.008, all the other tested SNPs’ genotypes were in Hardy-Weinberg equilibrium in the control group.
As presented in the [Table 4], the strongest significant association (a common OR=3.28) was evident between IGF2BP2 variant and T2D where the minor (T) allele and the TT genotype have a clear impact on the risk of disease in the patient group in an additive manner. To a lesser extent, the CDKNA2A/B polymorphic (T) allele polymorphism has a moderate effect on T2D development. A lower, but significant effect was also observed for COL8A1, KCNJ11, and KCNQ1 polymorphisms.
In COL8A1, IGF2BP2, CDKNA2A/B and KCNJ11, the minor allele and its homozygous genotype (when available) presented the risk-associated allele. In KCNQ1, however, the polymorphic allele seems to be protective.
Discussion
Genetic variation, in terms of SNPs in more than 80 loci are reported as risk alleles for T2D in various populations [5,6,24]. Variations in the genome- SNPs in particular-affect the level and function of gene expression and may modify the risk for having T2D. In this work, we could replicate the association between five documented SNPs and T2D in a Palestinian population. The significantly associated loci belong to one important aspect of T2D namely, pancreatic β-cell function/insulin secretion [7,25]. Additionally, the replication of those loci in Palestinian subjects further extends the trans-ethnic importance of many T2D genetic variants. The relatively small sample size employed presents one limitation of the current study and may reduce the chance of detecting the true effect of the investigated variants. Therefore, significant polymorphisms, in particular, need to be tested on a larger sample. On the other hand, selecting only middle age male subjects for the study makes the association between genetic variants and T2D more reliable.
Relevant to the pancreatic β-cell function/insulin secretion, our results showed that the genotype/allele frequencies of COL8A1 (rs792837 G>C), IGF2BP2 (rs4402960 G>T), CDKN2A/B (rs10811661 C>T), KCNJ11 (rs5219 C>T; E23K), and KCNQ1 (rs2237892 C>T) are significantly different between T2D patients and controls. Consistent with our results, significant association of these SNPs have been revealed also in other populations of diverse ancestries [26,32].
Despite their well-documented role in T2D in many genomewide association studies and meta analyses [24,33], TCF7L2 (rs793146), CDKAL1 (rs10946398), and SLC30A8 (rs13266634) did not show the same trend in our population. Still, lack of association of those polymorphism with T2D has been reported in some other populations [5,10,34,35].
Table 4: Genotype and allele frequencies and their effects on T2D risk for the 16 tested genes' polymorphisms.
Conflicting results are a common place in genetic association studies performed on different populations. Possible explanations for discrepant results include one or more of the following: differences in the ethnicity (genetic background), the sample size (i.e. statistical power), the characteristics of the study subjects (e.g. undefined chronic illnesses), presence of nucleotide polymorphism(s) somewhere else in the examined genes, epigenetic alterations, linkage disequilibrium to other sequence variants in the vicinity of the studied locus, and prevailing environmental conditions. It should be emphasized that frequency of T2D risk alleles and/or their effect size may be ethnicityrelated and in turn influence the detection of their association with T2D in a given population. Overall, T2D susceptibility variants may be categorized as common and ethnicity-specific that needs to beidentified for each population [5].
Conclusion
Results of the present study revealed that the five of investigated polymorphisms are significantly associated with T2D and could represent the first elements of “SNPs panel” for predicting T2D risk in the investigated population, particularly IGF2BP2. Future work should be directed towards testing those polymorphisms in T2D female patients, confirming the current findings with a larger sample, and examining additional SNPs that may help in characterizing additional genetic components of T2D.
Acknowledment
This study was supported by the Palestinian Medical Relief Society (PMRS) under the PRMS-NCD grant scheme (PRMS project BMZ-2014.1819.3).
References
- Iwata M, Maeda S, Kamura Y, Takano A, Kato H, et al. (2012) Genetic risk score constructed using 14 susceptibility alleles for type 2 diabetes is associated with the early onset of diabetes and may predict the future requirement of insulin injections among Japanese individuals. Diabetes Care 35: 1763-1770.
- Abu-Rmeileh N, Husseini A, O'Flaherty M, Shoaibi A, Capewell S, et al. (2012) Forecasting prevalence of type 2 diabetes mellitus in Palestinians to 2030: Validation of a predictive model. Lancet 300: S21.
- Dayeh TA, Olsson AH, Volkov P, Almgren P, Ronn T, et al. (2013) Identification of CpG-SNPs associated with type 2 diabetes and differential DNA methylation in human pancreatic islets. Diabetologia 56: 1036-1046.
- Sanghera DK, Blackett PR (2012) Type 2 diabetes genetics: beyond GWAS. J Diabetes Metab 3: 6948.
- Imamura M, Takahashi A, Yamauchi T, Hara K, Yasuda K, et al. (2016) Genome-wide association studies in the Japanese population identify seven novel loci for type 2 diabetes. Nat Commun 7: 10531.
- Lu J, Luo Y, Wang J, Hu C, Zhang R, et al. (2017) Association of type 2 diabetes susceptibility loci with peripheral nerve function in a Chinese population with diabetes. J Diabetes Investig 8: 115-120.
- Spellman CW (2010) Pathophysiology of type 2 diabetes: Targeting islet cell dysfunction. J Am Osteopath Assoc 110 (Suppl 2): S2-S7.
- Hertel JK, Johansson S, Midthjell K, Nygard O, Njoistad PR, et al. (2013) Type 2 diabetes genes-present status and data from Norwegian studies. Norsk Epidemiologi 23: 9-22.
- Li YY, Wang XM, Lu XZ (2014) KCNQ1 rs2237892 C→T gene polymorphism and type 2 diabetes mellitus in the Asian population: A meta-analysis of 15,736 patients. J Cell Mol Med 18: 274-282.
- Votsi C, Toufexis C, Michailidou K, Antoniades A, Skordis N, et al. (2017) Type 2 diabetes susceptibility in the Greek-Cypriot population: Replication of associations with TCF7L2, FTO, HHEX, SLC30A8 and IGF2BP2 polymorphisms. Genes (Basel) 8: E16.
- Frazer KA, Murray SS, Schork NJ, Topol EJ (2009) Human genetic variation and its contribution to complex traits. Nat Rev Genet 10: 241-51.
- Staiger H, Machicao F, Fritsche A, Häring HU (2009) Pathomechanisms of type 2 diabetes genes. Endocr Rev 30: 557-585.
- Yamaguchi S, Yamada Y, Matsuo H, Segawa T, Watanabe S, et al. (2007) Gender differences in the association of gene polymorphisms with type 2 diabetes mellitus. Int J Mol Med 19: 631-637.
- Kamura Y, Iwata M, Maeda S, Shinmura S, Koshimizu Y, et al. (2016) FTO gene polymorphism is associated with type 2 diabetes through its effect on increasing the maximum BMI in Japanese men. PLoS One 11: e0165523.
- Khan IA, Poornima S, Jahan P, Rao P, Hasan Q (2015) Type 2 diabetes mellitus and the association of candidate genes in Asian Indian Population from Hyderabad, India. J Clin Diagn Res 9: GC01-GC05.
- Yen CJ, Beamer BA, Negri C, Silver K, Brown KA, et al. (1997) Molecular scanning of the human peroxisome proliferator activated receptor gamma (hPPAR gamma) gene in diabetic Caucasians: identification of a Pro12Ala PPAR gamma 2 missense mutation. Biochem Biophys Res Commun 241: 270-274.
- Shaat N, Ekelund M, Lernmark A, Ivarsson S, Almgren P, et al. (2005) Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia 48: 2544-2551.
- Xu P, Che Y, Cao Y, Wu X, Sun H, et al. (2010) Polymorphisms of TCF7L2 and HHEX genes in Chinese women with polycystic ovary syndrome. J Assist Reprod Genet 27: 23-28.
- Huang Q, Yin JY, Dai XP, Pei Q, Dong M, et al. (2010) IGF2BP2 variations influence repaglinide response and risk of type 2 diabetes in Chinese population. Acta Pharmacol Sin 31: 709-717.
- Shabana, Hasnain S (2015) Effect of the common fat mass and obesity associated gene variants on obesity in Pakistani population: A case-control study. Biomed Res Int 2015: 852920.
- Xing G, Dongmei L, Yan W, Tao W, Wei L, et al. (2014) Correlation between KCNQ1 gene polymorphism and type 2 diabetes mellitus in Huaihai region of China. Fam Medi Community Health 2: 15-19.
- Singh S, Prasad SB, Yadav SS, Agrawal NK, Narayan G (2012) Association of common variants of CDKN2A/2B rs10811661 (C/T) and WFS1 rs6446482 (C/G) to type 2 diabetes mellitus in the Indian population of Eastern Uttar Pradesh. J Diabetes Metab 3: 227.
- Mansoori Y, Daraei A, Naghizadeh MM, Salehi R (2015) Significance of a common variant in the CDKAL1 gene with susceptibility to type 2 diabetes mellitus in Iranian population. Adv Biomed Res 4: 45.
- Prasad RB, Groop L (2015) Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 6: 87-123.
- Hardy OT, Czech MP, Corvera S (2012) What causes the insulin resistance underlying obesity? Curr Opin Endocrinol Diabetes Obes 19: 81-87.
- Almawi WY, Nemr R, Keleshian SH, Echtay A, Saldanha FL, et al. (2013) A replication study of 19 GWAS-validated type 2 diabetes at-risk variants in the Lebanese population. Diabetes Res Clin Pract 102: 117-122.
- Rao P, Wang H, Fang H, Gao Q, Zhang J, et al. (2016) Association between IGF2BP2 polymorphisms and type 2 diabetes mellitus: A case-control study and meta-analysis. Int J Environ Res Public Health 13: E574.
- Li H, Tang X, Liu Q, Wang Y (2013) Association between type 2 diabetes and rs10811661 polymorphism upstream of CDKN2A/B: a meta-analysis. Acta Diabetol 50: 657-662.
- Sokolova EA, Bondar IA, Shabelnikova OY, Pyankova OV, Filipenko ML (2015) Replication of KCNJ11 (p.E23K) and ABCC8 (p.S1369A) association in Russian diabetes mellitus 2 type cohort and meta-analysis. PLoS One 10: e0124662.
- Sun Q, Song K, Shen X, Cai Y (2012) The association between KCNQ1 gene polymorphism and type 2 diabetes risk: A meta-analysis. PLoS One 7: e48578.
- Kong X, Zhang X, Xing X, Zhang B, Hong J, et al. (2015) The association of type 2 diabetes loci identified in genome-wide association studies with metabolic syndrome and its components in a Chinese population with type 2 diabetes. PLoS One 10: e0143607.
- Peng S, Zhu Y, Xu F, Ren X, Li X, et al. (2011) FTO gene polymorphisms and obesity risk: A meta-analysis. BMC Med 9: 71.
- Diabetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, et al. (2014) Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 46: 234-244.
- Mtiraoui N, Turki A, Nemr R, Echtay A, Izzidi I, et al. (2012) Contribution of common variants of ENPP1, IGF2BP2, KCNJ11, MLXIPL, PPARY, SLC30A8 and TCF7L2 to the risk of type 2 diabetes in Lebanese and Tunisian Arabs. Diabetes Metab 38: 444-449.
- Florez JC, Jablonski KA, McAteer J, Sandhu MS, Wareham NJ, et al. (2008) Testing of diabetes-associated WFS1 polymorphisms in the diabetes prevention program. Diabetologia 51: 451-457.