Journal of Microbiology & Microbial Technology
Download PDF
Research Article
*Address for Correspondence: Rina Rani Ray, Post Graduate Department of Zoology, Bethune College, 181, Bidhan Sarani, Kolkata, West Bengal - 700006, India, E-mail: raypumicro@gmail.com
Citation: Ray RR, Debpali S, Kundu A. Preliminary Characterization of a NADPH Dependent Chromate Reductase from Trichoderma pseudokoningii. J Microbiol Microb Technol 2016;1(1): 4.
Copyright © 2016 Ray RR, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Microbiology & Microbial Technology | Volume: 1, Issue: 1
Submission: 24 May, 2016 | Accepted: 04 June, 2016 | Published: 08 June, 2016
Reviewed & Approved by: Dr. Aishwarya Devaraj, Center for Microbial Pathogenesis, Nationwide Children’s Hospital, Columbus, Ohio, USA
Preliminary Characterization of a NADPH Dependent Chromate Reductase from Trichoderma pseudokoningii
Rina Rani Ray*, Debpali Sur and Aditi Kundu
- Post Graduate Department of Zoology, Bethune College, Kolkata, West Bengal, India
*Address for Correspondence: Rina Rani Ray, Post Graduate Department of Zoology, Bethune College, 181, Bidhan Sarani, Kolkata, West Bengal - 700006, India, E-mail: raypumicro@gmail.com
Citation: Ray RR, Debpali S, Kundu A. Preliminary Characterization of a NADPH Dependent Chromate Reductase from Trichoderma pseudokoningii. J Microbiol Microb Technol 2016;1(1): 4.
Copyright © 2016 Ray RR, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Microbiology & Microbial Technology | Volume: 1, Issue: 1
Submission: 24 May, 2016 | Accepted: 04 June, 2016 | Published: 08 June, 2016
Reviewed & Approved by: Dr. Aishwarya Devaraj, Center for Microbial Pathogenesis, Nationwide Children’s Hospital, Columbus, Ohio, USA
Abstract
Intracellular chromate reductase enzyme extracted from a chromium tolerant strain Trichoderma pseudokoningii was partially purified by ultrafiltration and gel filtration chromatography. The purified fraction showed highest reducing activity in presence of 1% (w/v) hexavalent Chromium. NADPH at a conentration of 2 mM was found to act as electron donor in the reduction reaction. The intracellular chromate reductase was found to have an optimum pH and temperature of 7.4 and 70 °C respectively. The enhancement of reductase activity in presence of exogenous thiols indicated the presence of thiols at active site.Keywords
Trichoderma pseudokoningii; Chromate reductase; Hexavalent chromium; PurificationIntroduction
In nature, chromium exists in two oxidation states: hexavalent and trivalent, of which chromium (III) is considered to be one of the essential trace elements in the human body and is required in metabolism of glucose [1]. On the other hand, chromium (VI) is well known for its higher toxicity and carcinogenicity. Hence improper disposal of hexavalent chromium, discharged from tanneries, industries engaged in electroplating, paint pigment, dye manufacturing and stainless steel production makes chromium pollution one of the most important environmental problems in many regions of the world [2].Bioconversion of hexavalent chromium to its less toxic trivalent counterpart is the most promising way of bioremediation and is accomplished by enzyme chromium reductase. In aerobic conditions, the enzyme chromate reductase is reported to reduce Cr(VI) to Cr(III) inside or outside the plasma membrane and utilize NADH as electron donor, while under anaerobic condition membrane bound chromate reductase reduces Cr(VI) through respiratory chains involving cytochromes [3-5]. The most efficient Cr(VI) transforming enzymes that have been described to date are flavo-proteins, but these require expensive NADPH cofactors to power reduction, and stoichiometric addition of these cofactors is not an economically viable proposition for large scale applications [6-10]. Das and Chandra [11] have reported that Cr(VI) was reduced to Cr(III) in the presence of NADPH in the cell extract of Streptomyces species and existence of NADPH-dependent chromium (VI) reductase in Pseudomonas ambigua G-1, E.coli [12-13].
Many microorganisms have been reported to reduce the highly soluble and toxic Cr(VI) to the less soluble and less toxic Cr (III), of which fungi like Aspergillus niger and Aspergillus parasiticus [14], Paecilomyces [15], Fusarium sp. [16] and some yeasts are well known for their bio reduction potential [17].
Attempts have also been made to isolate the chromate reductase particularly from soluble cell-free extracts of Pseudomonas putida MK 1 but so far the literature survey is concerned, no such report is available on chromium reductase from Trichoderma spp. [12].
Present study includes isolation and partial purification chromate reductase enzyme from Trichoderma pseudokoningii and elucidation of its Cr(VI) reduction potential.
Materials and Methods
MicroorganismThe strain was isolated from the soil sample collected from tannery effluents in Kolkata, India and was screened by serial dilution for 3-4 times. The strain was identified as Trichoderma pseudokoningii by Agharkar Research Institute, Pune, India and was maintained at 4 °C for further studies [18].
Cultivation of the strain
For production of chromate reductase the strain was cultivated in 100 mL Erlenmeyer flasks each containing 10 mL Basal Medium (BM) composed of (g L-1): peptone 0.9; (NH4)2HPO4 0.4; KCl 0.1; MgSO4.7H2O 0.5%, dextrose and 0.2 mg ml-1 of K2Cr2O7 for 48 hours. The initial pH was adjusted to 7.0.
Enzyme extraction and purification
Cells mass were centrifuged at 8000 rpm for 7 min at 4 °C, suspended in phosphate buffer (10 mM, pH 7.4) and washed two times in the same buffer. Cells in ice bath were disrupted in a Sonicator (Rivotek, India) with an ultrasonic probe using two pulses of 20 s at 35 s each and the cell mass was extracted with 20 ml phosphate buffer (pH 7). The sonicated material of the fungal crude extract was centrifuged at 10,000 rpm at 4 °C for 10 min and filtered to obtain the soluble extract.
The enzyme extract was now ultrafiltered through a molecular membrane of 30 KDa cut off in a Tangential flow filtration system (PALL, Mumbai, India) and the concentrate was applied to a Superose 12 FPLC gel filtration column (50 X 20 cm) (Amersham Biosciences, Model No. 10/300 GL). The column was equilibrated with 10 mM phosphate buffer (pH 7) and fractions were collected by an automatic fraction collector at a flow rate of 1 mL/2 min. Each fraction was tested for chromium reductase activity and the fractions showing highest activity were collected, concentrated and used for further characterization.
Chromate reducing enzyme assay
The reaction mixture for the enzyme assay contained 500 μL of 1 mM of K2Cr2O7 as hexavalent chromium, 250 μL of the enzyme (purified fraction) and 250 μL of NADPH. The residual Cr(VI) was measured spectrophotometrically at 540 nm in UV visible spectrophotometer (Shimadzu, Japan) after 10 min using diphenylcarbazide method [19]. One unit of enzyme activity was defined as the amount of enzyme that reduced 1 nM of Cr (VI) per minute at 30 °C [20]. Proper controls were made one without the substrate and the other without the enzyme.
Protein measurement
The protein content of each fraction was measured at 280 nm and the total protein was measured by the Lowry method using bovine serum albumin (Sigma Chemical Co.) as standard [21].
Effect of pH and temperature
The optimum pH of the partially purified enzyme was determined by carrying out the chromate reduction at 70 °C at various pHs ranging from 6 to 8 maintained by 10 mM phosphate buffer. The optimum temperature of the partially purified enzyme was determined by incubating the reaction mixture of the enzyme at different temperatures ranging from 30 °C to 90 °C for 10 min. In order to determine the effects of pH on the stability of the enzyme, the partially purified enzyme protein was exposed to various pHs (6-8) for 30 minutes at 4 °C overnight. The thermostability of the enzyme was measured by exposing the enzyme protein at various temperatures (30 °C to 90 °C) maintaining the initial pH 7.4 for 30 minutes in water bath followed by rapid cooling.
Effect of electron donors and additives
Chromate reductase activity of the partially purified fraction of the enzyme was measured by incubating it in presence of various electron donors and additives (metal ions and thiol compounds) at a concentration of 1 mM and 10 mM respectively at 28 °C for 30 minutes followed by the assay of enzyme activity in usual procedure.
Atomic absorption spectroscopy
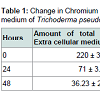
Change in extracellular and intracellular contents of chromium were determined by Atomic Absorption Spectrophotometer (Model- Perkin and Elmer 3110) analysed by UV spectrophotometer as per standard methods [22].
Chemicals
All reagents used were of analytical grade and were purchased from Sigma, USA, Himedia, India and Merck, Germany.
All experiments were done in triplicate and the values were averaged.
Results and Discussion
The working strain Trichoderma pseudokoningii was found to convert the hexavalent chromium and as evident from the data of atomic absorption spectroscopy (Table 1) it did not accumulate Cr(VI). Gradual and rapid depletion of hexavalent chromium from the extra and intra cellular medium indicated the reduction of Cr(VI) to Cr(III), which was accomplished by chromium reductase enzyme. Hence a chromium reductase enzyme must be synthesised by the strain.The fungal culture supernatant did not contain any reductase activity, whereas the pellet showed the presence of an active reductase enzyme having a specific activity of 1.03 U mg-1. The ultrafiltered chromate reductase (5 ml) subjected to gel filtration although showed four protein peaks at 280 nm, but gave a single peak of activity at 19th fraction (Figure 1). This fraction was subsequently used for elucidation of the properties of the enzyme.The partially purified enzyme showed temperature optima at 70 °C and was found to retain 71% of its activity at this temperature even after 10 minutes of exposure (Figure 2). Nearly similar temperature optimum of 80 °C was reported from Pseudomonas putida [20], whereas in Escherichia coli ATCC 33456 the chromium reductase showed maximum activity at 37 °C [13].
The chromium reductase enzyme was found to be active within a narrow range of pH and maximum activity was found to be at pH 7.4. The enzyme activity reduced sharply at a pH of 8 (Figure 3). This might be due to rapid denaturation of the enzyme protein at alkaline condition. Chromium reductase enzyme isolated from Escherichia coli ATCC 33456 showed maximum activity at pH 6.5 [13] whereas in Bacillus sp. ES29 (CRB) [19] and Pseudomonas G1DM21 [23] maximum enzymatic activity was found to be at neutral pH (pH 7).
The enzyme chromium reductase showed maximum activity (Figure 4) at 1% substrate concentration (1 mM of K2Cr2O7) and reduced most of the hexavalent chromium probably to its trivalent state within 15 minutes of incubation (data not shown). Very high concentration of substrate actually hindered the activity by saturating the total active site of the enzyme.
While studying the effect of various inhibitors on reductase activity, it was found that exogenous thiols like DTT, GSH and cysteine increased the enzymatic activity, which clearly indicated the presence of thiol residue(s) at the active site of the enzyme (Figure 5). Among the other metals and inhibitors Mg2+ acted as enhancer whereas heavy metals like Hg2+ and Cu2+ reduced the enzyme. Similar negative effect of metal ions like Hg2+, Zn2+, Ag2+, Cd2+ were also found in Escherichia coli ATCC 33456 [13].
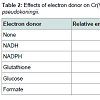
Among the electron donors tested, the reductase activity was found to increase upon supplementation of NADPH in the reaction mixtures (Table 2) the oxidation of which donates an electron to the chromate reductase enzyme, which in turn was transferred to Cr(VI), converting it to the intermediate form, Cr(V), which might have accepted two electrons from other organic substances to produce Cr(III) [17].
Conclusion
The present report deals with a preliminary study of the properties of a partially purified chromate reductase synthesised by a newly isolated strain of Trichoderma pseudokoningii. The enzyme was found to operate at high temperature and moderate alkaline condition in presence of NADPH as electron donor. The rapid rate of hexavalent chromium reduction efficacy of the enzyme extracted from the fungal strain might be used for effective bioremediation.References
- Inui T, Fujita K, Kitano M, Nakamura T (2010) Determination of Cr(III) and Cr(VI) at sub-ppb levels in water with solid-phase extraction/metal furnace atomic absorption spectrometry. Anal Sci 26: 1093-1098.
- Bartlett RJ, James BR (1996) Chromium. In: Sparks DL, Page AL, Helmke PA, Loeppert RH (Eds) Methods of soil analysis, Part 3, Chemical Methods. SSSA Book Series 5.3, SSSA, ASA, Madison, USA, pp. 683-701.
- Camargo FA, Bento FM, Okeke BC, Frankenberger WT (2003) Chromate reduction by chromium-resistant bacteria isolated from soils contaminated with dichromate. J Environ Qual 32: 1228-1233.
- Campos J, Martinez-Pacheco M, Cervantes C (1995) Hexavalent-chromium reduction by a chromate-resistant Bacillus sp. strain. Antonie Van Leeuwenhoek 68: 203-208.
- Dey S, Paul AK (2013) Evaluation of in vitro reduction of hexavalent chromium by cell-free extract of Arthrobacter sp. SUK 1201. Br Microbiol Res J 3: 325-338.
- Gonzalez CF, Ackerley DF, Park CH, Matin A (2003) A soluble flavoprotein contributes to chromate reduction and tolerance by Pseudomonas putida. Acta Biotechnol 23: 233-239.
- Kwak YH, Lee DS, Kim HB (2003) Vibrio harveyi nitroreductase is also a chromate reductase. Appl Environ Microbiol 69: 4390-4395.
- Opperman DJ, Piater LA, van Heerden E (2008) A novel chromate reductase from Thermus scotoductus SA-01 related to old yellow enzyme. J Bacteriol 190: 3076-3082.
- Opperman DJ, van Heerden E (2008) A membrane-associated protein with Cr(VI)-reducing activity from Thermus scotoductus SA-01. FEMS Microbiol Lett 280: 210-218.
- van der Donk WA, Zhao H (2003) Recent developments in pyridine nucleotide regeneration. Curr Opin Biotechnol 14: 421-426.
- Das S, Chandra AL (1990) Chromate reduction in Streptomyces. Experientia 46: 731-733.
- Suzuki T, Miyata N, Horitsu H, Kawai K, Takamizawa K, et al. (1992) NAD(P)H-dependent chromium (VI) reductase of Pseudomonas ambigua G-1: a Cr(V) intermediate is formed during the reduction of Cr(VI) to Cr(III). J Bacteriol 174: 5340-5345.
- Bae WC, Lee HK, Choe YC, Jahng DJ, Lee SH, et al. (2005) Purification and characterization of NADPH-dependent Cr(VI) reductase from Escherichia coli ATCC 33456. J Microbiol 43: 21-27.
- Shugaba A, Buba F, Kolo BG, Nok AJ, Ameh DA, et al. (2012) Uptake and reduction of hexavalent chromium by Aspergillus niger and Aspergillus parasiticus. Pet Environ Biotechnol 3: 1-8.
- C´ardenas-Gonz´alez JF, Acosta Rodr´ıguez I (2010) Hexavalent chromium removal by a Paecilomyces sp. fungal strain isolated from environment. Bioinorg Chem Appl 2010: 1-6.
- Sen M, Ghosh Dastidar M (2011) Biosorption of Cr (VI) by resting cells of Fusarium solani. Iran J Environ Health Sci Eng 8: 153-158.
- Arévalo-Rangel DL, Cárdenas-González JF, Martínez-Juárez VM, Acosta-Rodríguez I (2013) Hexavalent chromate reductase activity in cell free extracts of Penicillium sp. Bioinorg Chem Appl 2013: 1-6.
- Ray RR, Datta W, Sur D, Kundu A (2013) Optimization of fermentation parameters for the production of extracellular Endoglucanase, β-glucosidase and Endoxylanase by a chromium resistant strain of Trichoderma pseudokoningii. J Microbiol Biotechnol Food Sci 3: 54-58.
- Conceição DP, dos Passos CT, Jacques RJ, Bento FM, Simonetti AB, et al. (2009) A novel chromate reductase from Bacillus sp. ES29: characterization and partial purification. Revista Ciências Exatas e Naturais 11: 237-256.
- Park CH, Keyhan B, Wielinga B, Fendorf S, Matin A (2000) Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl Environ Microbiol 66: 1788-1795.
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265-275.
- Page AL, Miller RH, Keeney DR (1982) Methods of soil analysis. Part 2, Chemical and microbiological properties, American Society of Agronomy, Soil Science Society of America Madison, Wisconsin, USA.
- Desai C, Jain K, Madamwar D (2008) Hexavalent chromate reductase activity in cytosolic fractions of Pseudomonas sp. G1DM21 isolated from Cr(VI) contaminated industrial landfill. Process Biochem 43: 713-721.