Journal of Forensic Investigation
Download PDF
Case Report
*Address for Correspondence: M Lurdes Pontes, Instituto Nacional de Medicina Legal e Ciências Forenses, Portugal, E-mail: mlurdes.pontesrebelo@gmail.com
Citation: Pontes ML, Amigo OM, Fondevila M, Porto MJ, Medeiros R. Analysis of Autosomal SNPs in Paraffin-embedded Tissue Samples: Two Cases. J Forensic Investigation. 2015;3(1): 4.
Copyright © 2015 Pontes ML, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Forensic Investigation | ISSN: 2330-0396 | Volume: 3, Issue: 1
Submission: 13 January, 2015| Accepted: 07 February, 2015 | Published: 11 February, 2015
Analysis of Autosomal SNPs in Paraffin-embedded Tissue Samples: Two Cases
M Lurdes Pontes1,2*, Olalla Maroñas Amigo3, Manuel Fondevila3, M. João Porto1,2 and R. Medeiros4,5,6
- 1Instituto Nacional de Medicina Legal e Ciências Forenses, Portugal
- 2CENCIFOR- Forensic Science Center, Portugal
- 3Institute of Legal Medicine, Universidade de Santiago de Compostela, Santiago de Compostela, Spain
- 4Molecular Oncology group, Portuguese Institute of Oncology, Portugal
- 5ICBAS, Abel Salazar Institute for the Biomedical Sciences, University of Porto, Largo Professor Abel Salazar, 4099-003, Porto, Portugal
- 6LPCC, Research Department-Portuguese League Against Cancer (NRNorte), Estrada Interior da Circunvalação 6657, 4200 Porto, Portugal
*Address for Correspondence: M Lurdes Pontes, Instituto Nacional de Medicina Legal e Ciências Forenses, Portugal, E-mail: mlurdes.pontesrebelo@gmail.com
Citation: Pontes ML, Amigo OM, Fondevila M, Porto MJ, Medeiros R. Analysis of Autosomal SNPs in Paraffin-embedded Tissue Samples: Two Cases. J Forensic Investigation. 2015;3(1): 4.
Copyright © 2015 Pontes ML, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Forensic Investigation | ISSN: 2330-0396 | Volume: 3, Issue: 1
Submission: 13 January, 2015| Accepted: 07 February, 2015 | Published: 11 February, 2015
Abstract
Despite the numerous STR loci available for analysis of cases submitted to forensic laboratories, namely with the use of commercial kits that amplify mini - short tandem repeats (STRs), some cases remain “unsolved” i.e. fail to yield a STR DNA profiling result or result in likelihood ratio calculations that are not sufficiently high. Autosomal single nucleotide polymorphisms (SNPs) may constitute a potential advantage to solve some of these difficult cases, as has already been described by others.In this work we present two revisited cases of our routine work, where the type of samples available for analysis was histological blocks of paraffin-embedded tissue samples from a man (Case 1-“lymph node”) and from a woman (Case 2). The application of autosomal SNPs in the analysis of paraffin-embedded tissues constitutes one of the possible approaches that can be used in our laboratory in some “difficult” cases, and is part of a validation process of the described methodology.
Following extraction the two sample DNAs had already been genotyped with the routine commercial STR or mini-STR kits, like AmpFLSTR® Identifiler®, MiniFilerTM and NGMTM PCR Amplification Kits and compared to their respective reference samples. These samples were then typed for the 52 loci included in the in the SNP for ID 52 plex using a SNaPshotTM assay previously described by Sanchez et al. in order to test and validate this multiplex in real casework [1].
The study showed that autosomal SNP can be used as additional markers to the profiling of difficult samples such as paraffin embedded tissue.
Keywords
Forensic genetics; Autosomal single nucleotide polymorphisms; 52-SNP-multiplex; Casework; Paraffin-embedded tissue samplesIntroduction
Currently forensic laboratories solve almost all the requested cases through the traditional use of commercial kits that amplify short tandem repeats (STRs) or mini-STRs. In spite of this, there are still some “unsolved”or challenging cases that are occasionally encountered in forensic laboratories like paraffin-embedded tissue. “Unsolved” or challenging cases can be paternity cases, usually where the father is deceased and other relatives are disposable to investigate the paternity, that often lead to low paternity indexes after STR profiling. Paraffin embedded samples are not usual in forensic investigations. But these types of samples due to their high level of degraded sample DNAs, may also lead to low Likelihood Ratios (LR) after conventional STR analysis.The use of autosomal SNPs for forensic research has been widely discussed in recent years. SNPs have important advantages compared to short tandem repeats (STRs) [2] and in combination with standard markers, improve the power of discrimination and provide potential supplementary data for paternity testing [3,4]. Moreover their study may constitute an improvement in the analysis of degraded samples [5].
As it was already said, paraffin embedded samples are not usual in forensic investigations. Here, the only two interesting cases involving paraffin-embedded tissue samples we have had over the years are presented. These cases were previously resolved using the traditional STR markers, but they were revisited in order to access the relevance of studying autosomal SNPs. This was achieved by calculating the LR before and after adding the information of the analyzed SNPs. The use of SNPs for challenging forensic sample type’s isdocumented by others, as is the recovery and analysis of DNA from formalin-fixed paraffin-embedded (FFPE) tissues but mostly in medical context and using other analysis methodologies. Equally, the combining of statistical data from STR and SNP loci has been demonstrated before.
In spite of these two cases the STR results alone would have provided sufficient certainty of source identification to be used without the additional SNP work, our aim was to validate the use of the SNP for ID 52 plex and demonstrate the increment of the calculated LRs after their use as additional markers to the profiling of difficult samples such as paraffin embedded tissue, in order to apply their use in similar cases that may be encountered in our forensic laboratory in the future.
Materials and Methods
SamplesIn this study histological material from two persons with a histological tumor - positive diagnostic was available. The aim of the requested examinations was to confirm the identity of the persons involved, supposedly the donors of the paraffin-embedded samples, who questioned the results of the histology laboratory biopsies observations. For that, a DNA profile had to be obtained from cuts of the histological blocks of paraffin-embedded tissue samples (Figure 1); then it was necessary to compare the obtained profiles with the ones of the reference samples from the donors of the histological samples, respectively a man and a woman. Extraction
DNA of reference samples (buccal swabs) was extracted using the slightly modified Chelex® method [6] and the DNA from the casework samples was extracted using the EZ1 DNA Tissue Kit together with the EZ1 DNA Forensic Card protocols with a BioRobot® EZ1 (Qiagen) [7].
Quantification
DNA concentrations were determined by real-time PCR using the QuantifilerTM Human DNA Quantification Kit (AB) and the ABI PRISM® 7500 Sequence Detection System (AB). The final concentrations of the casework samples were 0.02 and 0.05 ng/μl, respectively.
PCR amplification
The STR and mini-STR amplification of the extracted samples was achieved following the manufacturer protocols for the kits Identifiler® and MinifilerTM or NGMTM respectively.
A total of 52 autosomal SNPs were amplified by polymerase chain reaction (PCR) in 12.5 μl reactions containing DNA, in the case of these particular casework samples we adjusted the final volume to 25 μl in order to have a higher input of DNA sample. The PCR conditions were performed as previously described [1]. When using the problem samples of cases 1 and 2, we used a DNA input of ~0.10 and ~0.25 ng respectively.
SBE reaction
Two single base extension (SBE) reactions were performed (Auto 1 and Auto 2) as previously described with slight modifications [1] and included 2.5 μl SNaPshot reaction mix (Applied Biosystems - AB), 1.5 μl SBE primer mix (0.01–0.27 μM) and 2 μl of purified PCR product (with Exonuclease I/Shrimp Alkaline Phosphatase - ExoIFigure SAP), totaling 6 μl. Excess nucleotides were then removed by the addition of 1 μl SAP (1 U/μl) to the SBE mix. The multiplexes were balanced in the laboratory in order to validate the described method in our particular conditions.
Typing
STR and SBE products separation was performed by capillary electrophoresis, using an ABI Prism 3130 xl Genetic Analyzer (AB), with 36 cm capillary arrays and POP-4 polymer (Applied Biosystems, Foster City, CA, USA). Analysis was made using GeneMapper® ID-X. For the autosomal SNPs allele calls were made manually.
Quality control
The 52 plex SNP assay was validated, locally and through interlaboratory collaborative exercises, promoted by the GHEP-ISFG group.
Analysis of data
The LRs were calculated using an in house excel spread sheet, simply by multiplying the individual LRs from each of the SNP markers, using the previous published frequencies for those markers in a population sample of the North of Portugal, assuming that all the markers are independent [8].
Results
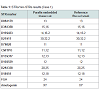
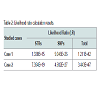
The results obtained for the STRs for Case 1 are presented in Table 1. These were only possible for 11 and 14 STR markers, respectively for case 1 and 2. Concerning SNPs, as it was expected, the results were possible for all the 52 typed SNPs in one of the cases (Case 1) and for 50 SNPs in the other (Case 2). The results for Case 2 are shown (Figure 2).The impact of studying SNPs in the overall Likelihood Ratio (LR) is presented by calculation of this parameter before and after adding the additional information given by the typed SNPs (Table 2), assuming all the studied loci are independent. Combining of statistical data from STR and SNP loci has been demonstrated before, but it is still not known if all SNP loci are independent of each other or of the STR loci used in this study. It is only known that this set of 52 SNP markers was carefully tailored in order to be sufficiently apart from each other guaranteeing this independence as reported by Sanchez et al. [1].Before this example of autosomal SNP application some validation work was made in our laboratory, namely using internal controls of known standard DNAs and through inter-laboratory collaborative exercises, promoted by the GHEP-ISFG group. We also used allele frequencies for the 52-SNPs on our reference population – north of Portugal population [8].
Discussion and Conclusion
The novelty of this work consisted in the application of the 52-SNP multiplex previously described [1] for study paraffin-embedded tissue samples, traditionally not usual in forensic cases. In the medical field there are many studies that involve paraffin-embedded samples. Another study looked at paraffin-embedded tissue for identification of the tissue source using a SNP multiplex, but with a more recent methodology [9].The typing results and the performed LR calculations revealed that the 52 plex SNP assay may constitute a robust and sufficiently sensitive tool to complement the results from the traditional markers (STR and mini-STRs) in difficult forensic casework, which involves samples with material particularly degraded, like paraffin-embedded tissue samples, old cadaveric remains like bones as it was already reported [10], and other low-template DNA and highly degraded DNA from crime case samples involved in routine casework [11,12].
Acknowledgements
We thank Prof. María Victoria Lareu from the Institute of Legal Medicine of the University of Santiago de Compostela, Galicia, Spain, for her support and advising.References
- Sanchez JJ, Phillips C, Børsting C, Balogh K, Bogus M, et al. (2006) A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis 27: 1713-1724.
- Gill P (2001) An assessment of the utility of single nucleotide polymorphisms (SNPs) for forensic purposes. Int J Legal Med 114: 204-210.
- Amorim A, Pereira L (2005) Pros and cons in the use of SNPs in forensic kinship investigation: a comparative analysis with STRs. Forensic Sci Int 150: 17-21.
- Phillips C, Fondevila M, García-Magariños M, Rodriguez A, Salas A, et al. (2008) Resolving relationship testes that show ambiguous STR results using autosomal SNPs as supplementary markers. Forensic Sci Int Genet 2: 198-204.
- Butler JM, Coble MD, Vallone PM (2007) STRs vs. SNPs: thoughts on the future of forensic DNA testing. Forensic Sci Med Pathol 3: 200-205.
- Walsh PS, Metzer DA, Higuchi R (1991) CHELEX 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 54: 134-139.
- (2004) EZ1 DNA Handbook, Qiagen.
- Lurdes Pontes M, Pinheiro MF (2014) Autosomal SNPs study of a population sample from North of Portugal and a sample of immigrants from the Eastern Europe living in Portugal. Leg Med (Tokyo) 16: 118-120.
- Miller JK, Buchner N, Timms L, Tam S, Luo X, et al. (2014) Use of Sequenom sample ID Plus® SNP genotyping in identification of FFPE tumor samples. PLoS One 9: e88163.
- Fondevila M, Phillips C, Naveran N, Fernandez L, Cerezo M, et al. (2008) Case report: Identification of skeletal remains using short-amplicon marker analysis of severely degraded DNA extracted from a decomposed and charred femur. Forensic Sci Int Genet 2: 212-218.
- Børsting C, Mogensen HS, Morling N (2013) Forensic genetic SNP typing of low-template DNA and highly degraded DNA from crime case samples. Forensic Sci Int Genet 7: 345-352.
- Freire-Aradas A, Fondevila M, Kriegel AK, PhillipsC, Gill P, et al. (2012) A new SNP assay for identification of highly degraded human DNA. Forensic Sci Int Genet 6: 341-349.