Journal of Veterinary Science & Medicine
Download PDF
Research Article
Characterisation and Recognition by Immune Hosts of a Sheep Nematode Parasite Teladorsagia circumcincta Chitinase
Umair S*, Bouchet C and Baten A
AgResearch Ltd, Hopkirk Research Institute, Grasslands Research Centre, New Zealand
*Address for correspondence: Umair S, AgResearch Ltd, Hopkirk Research Institute, Grasslands Research
Centre, Private Bag 11-008, Palmerston North 4442, New Zealand,
Email: saleh.umair@agresearch.co.nz
Submission: 8 June, 2021;
Accepted: 2 August, 2021;
Published: 5 August, 2021
Copyright: © 2021 Umair S et al. This is an open access article
distributed under the Creative Commons Attribution License, which
permits unrestricted use, distribution, and reproduction in any medium,
provided the original work is properly cited.
Abstract
A 912 bp full length cDNA encoding Teladorsagia circumcincta
chitinase (TciCHT) was cloned and expressed in Escherichia coli.
Recombinant TciCHT was purified and its enzyme assays performed.
The predicted protein consisted of 304 amino acids and weighed
about 34 kDa on sodium dodecyl (lauryl) sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE). The recombinant TciCHT was expressed
as inclusion bodies and treated with 8M urea to denature the
protein. Multiple alignments of the protein sequence of TciCHT with
homologues from other helminths showed that the highest similarity
(88%) to the CHT of Haemonchus sp, and 65-87% similarity to the other
nematode CHT. Substrate binding sites and conserved regions were
identified and shown to be conserved in other homologues. Enzyme
assays were carried out using multiple substrates but failed to produce
any activity. Recombinant TciCHT was recognised by antibodies in
both serum and saliva from field-immune sheep in ELISA, however, that
was not the case with nematode-naïve sheep. Given the importance
of the enzyme and its recognition by the immune-sheep, Teladorsagia
circumcincta chitinase might have potential as a vaccine candidate
to control this common sheep parasite.
Introduction
Teladorsagia circumcincta is a mucosal browser and resides in the
abomasa of the ruminants. The parasite has a direct life-cycle where
the eggs laid by the adult worms are passed on to the pasture through
faeces and eggs develop into the infective stage larvae (L3), which are
ingested and reside in the abomasa of the ruminants and develop into
adult worms. Parasitic nematode worm infection is one of the biggest
health problems for farmed ruminants worldwide. Parasitic worm
infections are harmful to a host animal for many reasons and cause
costly production losses and if left untreated, animals can die causing
further economic loss to farmers.
The control and productivity losses caused by parasitic nematodes
cost the New Zealand livestock industry ~$700 million annually.
Currently, farmers rely on the use of anthelmintics to control parasitic
nematodes, however resistance of parasites to one or more of these
agents is now widespread. Recent industry-funded surveys in New
Zealand found that 64% of sheep farms and 94% of beef farms now
have parasites that are resistant to at least one of the anthelmintics
[1]. It is really important to understand worm biology and look for
the targets that are essential for the worm survival.
Chitinase catalyses the hydrolytic cleavage of the ß-1, 4-glycoside
bonds present in biopolymers of N-acetylglucosamine, particularly
in chitin. Chitinases are widely distributed in living organisms
and are found in fungi, bacteria, parasites, plants and animals. The
chitinolytic enzymes are also characterised based in their enzymatic
action on chitin substrates. Endochitinases are the enzymes catalysing the random cleavage at internal points in the chitin chain whereas
exochitinases catalyse the progressive release of acetylchitobiose from
the non-reducing end of chitin.
Chitinases perform a variety of function depending on the
organism they are present in, for example, in bacteria chitinases
are mainly involved in the nutritional processes [2,3] whereas in
yeast and fungi, these enzymes participate in morphogenesis [4,5].
In animals and plants, chitinases primarily play a role to defend the
organism against infections [6-8] and regulate innate immunity and
tissue function. In parasites, chitinase is a significant component of
the eggshell and play a vital role in egg hatching [9].
Chitin is one of the most abundant polysaccharide and in
nematode parasite comprises cuticles, egg-shell, pharynx and microfilarial
sheath[10-12]. Nematode chitinases consists of multiple
genes and it is believed that the enzymes have additional roles in the
nematode life cycle because of the presence of stage-specific gene
expression Chitinase was detected in the excretory/secretory (ES)
protein of the root-knot nematode[13]. Helminth chitinase are
induced during T helper- type responses and contribute to asthma,
fibrosis and helminth immunity [14,15]. The chitin metabolism
can provide unique targets for parasite control because chitin is
not found in vertebrates. Because of being central in the chitin
metabolism, chitinase has potential as a vaccine candidate. Mice
vaccinated with chitinase DNA resulted in partial protection against
Onchocerca volvulus [16]. Similarly, rabbits and mice vaccinated with
recombinant chitinase-like proteins provided protection against mite
Sacroptes scabiei and hard tick Haemaphysalis longicornis respectively
[17,18].
The aim of the present experiments was to determine the fulllength
sequence of chitinase in the sheep abomasal nematode parasite
Teladorsagia circumcincta, express the protein, structural analysis
and antigenicity of the recombinant protein.
Materials and Methods
All chemicals used in these experiments were purchased from
the Sigma Chemical Co. (Mo, USA) unless stated. Use of lambs for
parasite culturing and harvesting adult worms for molecular biology
studies has been approved in protocol # 13502 by the AgResearch
Grasslands Animal Ethics Committee
Parasite culture and collection:
Pure cultures of T. circumcincta were obtained by passaging larvae
through sheep and adult worms recovered as described previously
[19].RNA isolation and cDNA synthesis:
RNA was isolated from adult worms and first strand cDNA
synthesised from 1 ug total RNA using the iScript Select cDNA
Synthesis Kit (Bio-Rad) as as described previously [19].Cloning and expression of T. circumcincta recombinant TciCHT in E. coli:
A partial T. circumcincta chitinase sequence (TELCIR_01681)
was obtained from AgResearch’s internal database. In order to obtain
the full length chitinase gene sequence, Rapid Amplification of cDNA
Ends (RACE) using the SMARTer RACE cDNA Amplification Kit
(Clontech) was carried out. Both 3’ end and 5’ RACE primers were
designed but failed to get a full-length gene. Nested PCR reactions
were carried out using SL1 and SL2 primers to detect the presence of
longer transcripts. The gene sequence was sent to GenScript (Hong
Kong) for gene synthesis and insertion into PUC57. The TciCHT
gene were cloned, using protocols described previously [20] into the
expression vector AY2.4. Restriction enzymes Ndel and Notl were
used in cloning and the resulting protein was N-terminal His tagged
recombinant.E. coli strain BL21 (DE3) transformed with AY2.4 TciCHT as
described previously [21] was grown in Luria Broth (LB). L-Arabinose
was added as inducer and the culture grown for an additional 2h at
30 oC and 250rpm. Bacteria were harvested as described before.
Briefly, The pellet was weighted and resuspended in 10ml per gram
of pelleted bacteria of equilibration buffer (20mM sodium biphosphate,
0.5M NaCl, 20mM Imidazole, pH 7.4). Protease inhibitors
were added to the suspension, which was then passed through, the
chamber of a MP110 Microfluidizer® (Microfluidics, USA) seven
times consecutively under ice, at 20,000psi to ensure the full lysis of
E.coli as recommended by the manufacturer. The crude lysate was
centrifuged at 15,000g for 20min at 4oC to remove all cell debris and
the supernatant collected and filtered through a 0.22μm to insure the
removal of further impurities.
Recombinant TciCHT was expressed as totally insoluble protein
and the protein was purified and folded as inclusion bodies as
described [22]. Briefly, purified recombinant poly-histidine TciCHT
was obtained by Fast protein liquid chromatography (FPLC) under
native conditions using from a Ni-NTA column (Qiagen), completed
with a BIO-RAD chromatography system (Bio-Rad, USA). Sodium
bi-phosphate buffer was used as an equilibration buffer, sodium biphosphate
containing 20mM imidazole and 8M urea as the wash
buffer and sodium bi-phosphate containing 500mM imidazole and
8M urea as elution buffer. The recombinant TciCHT was purified
as inclusion bodies, therefore, dialysed in the buffer containing 8M,
6M, 4M, 2M and no urea for 12h in each buffer at 4oC. The protein
concentration was determined by the Nanodrop (A280nm assay)
using extinction coefficient (92610M-1cm-1) and molecular weight
(34KDa)
Purification and gel Electrophoresis:
Recombinant TciCHT was produced as recombinant polyhistidine
protein and was obtained by FPLC under native conditions
using a Ni-NTA column (Qiagen), and a Biologic DUO-FLOW BIORAD
chromatography system (Bio-Rad, USA) as described before
[22].Protein Structure Modelling:
CHT sequences from several closely related helminth species
including H. contortus were collected from NCBI. Protein alignments
were performed using the Muscle multiple alignment option in
Geneious 8 (Biomatters Ltd) with the Blosum 62 similarity matrix.
Iterative threading assembly refinement (I-TASSER) [23], was used to
construct a structural model of TeciCHT. For each target, I-TASSER
simulations generate a large ensemble of structural conformations,
called decoys. The confidence of each model is quantitatively
measured by the C-score that is calculated based on the significance
of threading template alignments and the convergence parameters of
the structure assembly simulations. Another important metric is TMscore
which is estimated from C-score and is used for measuring the
similarity of two protein structures/ Scores higher than 0.5 assumes
the parent structure and modelled protein share the same fold while
below 0.17 suggests a random nature to the produced model.Chitinase assay:
Recombinant chitinase activity was measured as described.
The kit contains three different substrates to measure endo- and
exo-chitinase activity. Each substrate was dissolved in DMSO and
diluted 1:20 in the assay buffer (100 mM citric acid, 200 mM sodium
phosphate, pH 5.5) prior to the assay. The assay started after the
addition of recombinant TciCHT and incubated for 30 min at 37 ˚C.
The reaction was stopped by the addition of stop solution (500mM
sodium carbonate, 500 mM sodium bicarbonate, pH 10.5) and the
liberated 4-MU was measured at 450nm.Host Recognition:
To test for the presence of antibodies in the blood and saliva that
react with the recombinant enzyme, saliva and serum samples taken
from parasite-exposed and -naïve sheep as described previously. The
pooled serum and saliva samples used for ELISA were collected from
18 male Romney lambs 6-7 months-old and previously exposed in the
field to multiple species of parasites and had developed immunity to
T. circumcincta infection. TciCHT (5 μg/ml) was immobilised onto
ELISA plates (Maxisorp, Thermo Scientific) overnight. Free binding
sites were then blocked with Superblock (Thermo Scientific, USA) and
then incubated with serial dilutions (200- to 6400-fold for serum or
20- to 160-fold for saliva) in ELISA buffer for 2h at room temperature.
Bound serum immunoglobulins were then detected with rabbit antisheep
Ig-HRP (Dako, Denmark), diluted 1:5000 by incubation for 1h
at 37 °C and the colour developed with 3,3’,5,5’ tetramethylbenzidine
(AppliChem, Germany). Saliva IgA was detected with rabbit antisheep
IgA-HRP, which was diluted, incubated and the colour
developed, as described for serum Ig.Results
TciCHT gene sequence and structure:
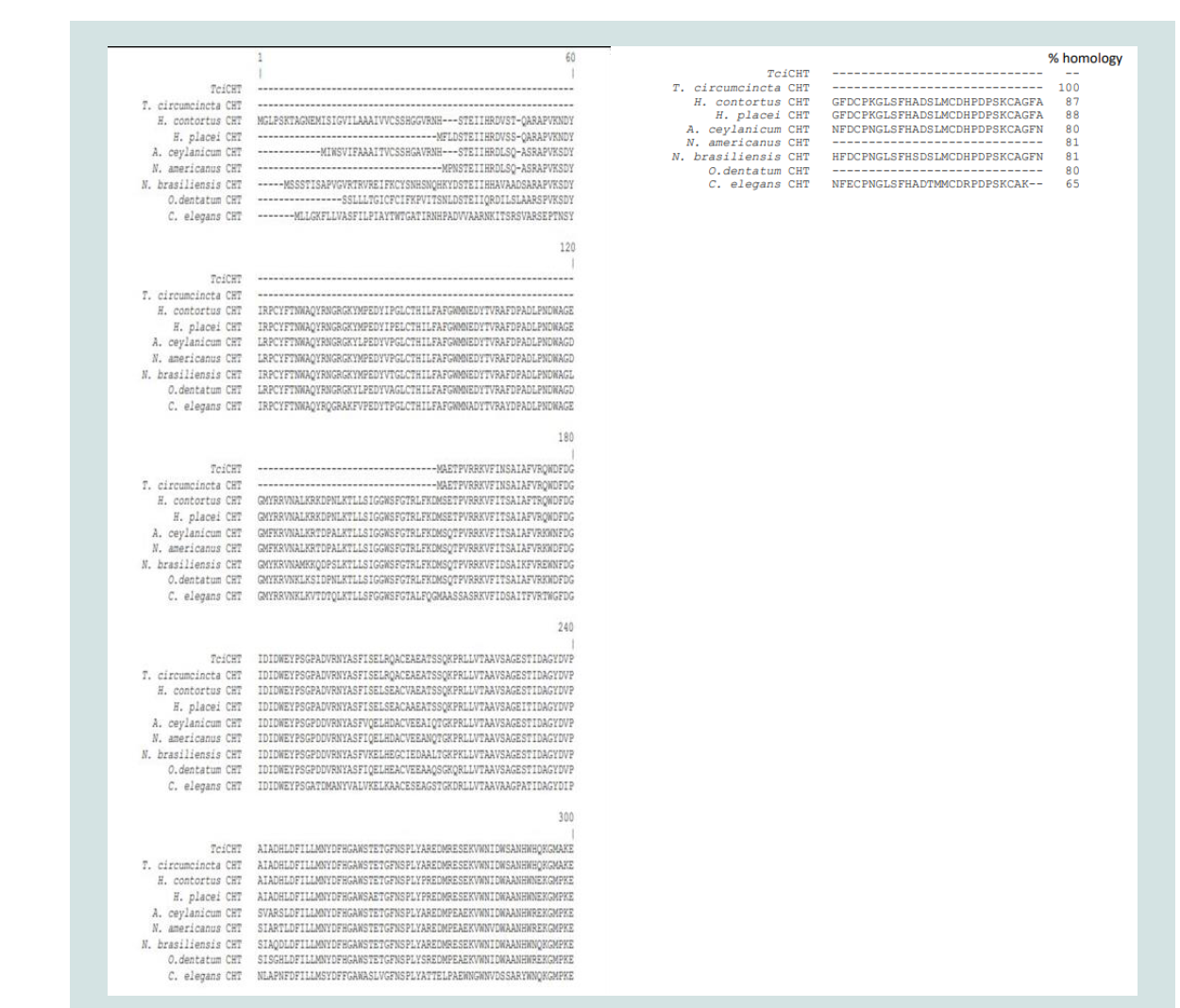
The 912 bp full length T. circumcincta cDNA sequence has been deposited in Genbank as Accession No. KX452945. The predicted protein consisted of 314 amino acids (Figure 1). Multiple alignments,
using Alignment Geneious 8, of the protein sequences of TciCHT
with homologues from published T. circumcincta, H. contortus, H.
placei, A ceylanicum, N. americanus, N. brasiliensis, O. dentatum and
C. elegansare shown in Figure 1.(Figure 1). Multiple sequence alignment of TciCHT with
homologues from H. contortus (GI: CDJ82138), H. placei (GI:
VDO51030), A. ceylanicum (GI: EYC03522), N. americanus (GI:
XP013294292), N. brasiliensis (GI: VDL62424), O. dentatum (GI:
KHJ90958), C. elegans (GI: NP508588) homologues. Amino acid
residues indicated in bold are essential to the enzyme activity.
Figure 1: Multiple sequence alignment of TciCHT with homologues from H. contortus (GI: CDJ82138), H. placei (GI: VDO51030), A. ceylanicum (GI:
EYC03522), N. americanus (GI: XP013294292), N. brasiliensis (GI: VDL62424), O. dentatum (GI: KHJ90958), C. elegans (GI: NP508588) homologues. Amino
acid residues indicated in bold are essential to the enzyme activity.
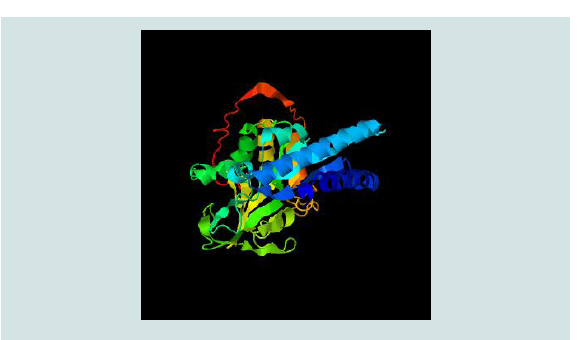
The predicted 3D structure of TciCHT is shown in Figure 2. It
has the highest structural similarity with 5WUP which is insect group
III chitinase (CAD1) from Ostrinia furnacalis [24]. The best model
predicted by I-TASSER has a C-score 1.36 and a TM-score of 0.90 ±
0.06 which indicates high quality of the prediction. (Figure 2) shows the
model quality in terms of Z-score by ProSA-web. The plot shows the
Z-scores of all experimentally determined protein chains in current
PDB and the position of the predicted TciCHT model
Figure 2: The predicted tertiary structure of TciCHT. Coiled ribbons represent
alpha helix whereas flat ribbons beta sheets whereas colours represent
protein subunits.
Figure 2. The predicted tertiary structure of TciCHT. Coiled
ribbons represent alpha helix whereas flat ribbons beta sheets whereas
colours represent protein subunits.
Recombinant protein expression:
A number of varying conditions were used in the trial expression
and based on which maximal production of functional recombinant
TciCHT was obtained in the E. coli strain BL21 (DE3) when expression
was induced with 0.2% L-arabinose for 3h at 37 ºC. The recombinant
TciCHT was expressed as inclusion bodies and the inclusion bodies
were treated using urea. The protein was dialysed in decreasing urea
concentrations to facilitate the folding. Recombinant TciCHT was
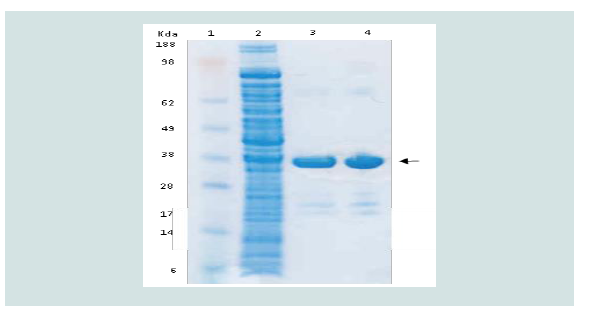
purified as N-terminal His-tagged protein with weight of about 34
kDa (Figure 3). The presence and the purity of recombinant TciCHT
were confirmed by Western blotting.
Figure 3: Purified recombinant TciCHT on Bis-Tris protein gel stained with
SimplyBlue safe stain. Lane 1: Standards in KDa; Lane 2: Filtered soluble
lysate; Lane 3: recombinant TciCHT; Lane 4: recombinant TciCHT after
dialysis and refolding.
TciCHT assay:
No enzyme activity was detected with either substrate over pH
6-10 or enzyme concentration between 50-250 μg.Host recognition:
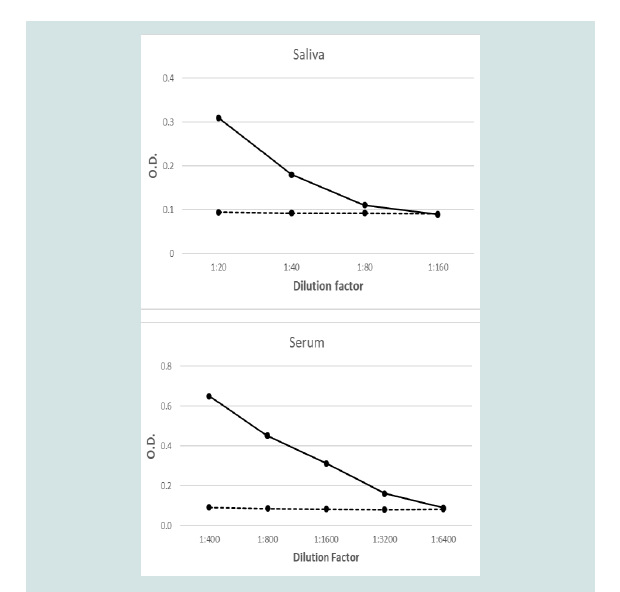
Recombinant TciCHT was recognised in by antibodies in saliva
and serum samples from the parasite-exposed animals whereas that
was not the case of the samples collected from parasite-naïve animals
(Figure 4).Discussion
This study showed the close relationship between a T. circumcincta
chitinase (TciCHT) to that from other helminth homologues. TciCHT gene, which encoded 912 bp was cloned, expressed in E. coli.
Recombinant TciCHT consisted of 30 4 amino acid and the protein
was recognised by antibodies using ELISA in the saliva and serum
from the sheep that were immune to parasites, but not nematodenaïve
animals.
A 912 bp full length cDNA sequence encoding T. circumcincta
chitinase (TciCHT) was amplified from adult T. circumcincta cDNA,
cloned and expressed in E. coli. The 304 amino acid TciCHT protein
expressed in E. coli was typical of CHT identified and characterized
in several helminths. The TciCHT protein had 87% homology to
H. contortus homologue and 65-80% similarity to other nematode
and trematode homologues (Figure 1). Our analysis showed a high
similarity of the TciCHT protein with CHT form other closely related
species. We identified the most likely TciCHT model and the analysis
revealed high similarity with insect group III chitinase (CAD1) from
Ostrinia furnacalis.
No enzyme activity was detected with either of the three
substrates or varying pH or enzyme concentrations. The recombinant
TciCHT protein was expressed in E. coli as inclusion bodies and
lacked biological activity. Proteins expressed as inclusion bodies may
not be folded correctly and be functional enzymes [26-28]. Although,
the TciCHT sequence appears to be truncated with both 3’ and 5’
ends incomplete, his doesn’t seem to be the case as both 3’ and 5’
RACE was performed, which failed to detect any longer sequence
(Figure 1). It is interesting to note that TciCHT fully matched with
the sequence available in the database indicating the possibility of a
shorter sequence compared to other helminth CHT sequences.
Previous studies on the potential of fungal chitinase have shown
very promising results and the enzyme has captured and completely
destroyed nematode eggs [29] and animals vaccinated with a recombinant chitinase resulted in high levels of protection [30-32].
Native T. circumcincta CHT is part of worm’s ES products, highly
antigenic and antibodies in both serum and saliva from field-immune
sheep recognised recombinant TciCHT in an ELISA (Figure 4). These
findings are very promising and further studies will validate the
protective efficacy of recombinant TciCHT.
Summary
A 912 bp cDNA encoding Teladorsagia circumcincta chitinase was
cloned and expressed in Escherichia coli. Multiple alignments of the
protein sequence of TciCHT with homologues from other helminths
showed good similarity to the other helminth CHT. Recombinant
TciCHT was recognised by antibodies in both serum and saliva from
field-immune sheep in ELISA.
Acknowledgments
The financial support of AGMARDT (Grant No. P14003) is gratefully acknowledged