Journal of Veterinary Science & Medicine
Download PDF
Research Article
*Address for Correspondence: Huaguang Lu, Animal Diagnostic Laboratory, Department of Veterinary and Biomedical Science, Pennsylvania State University, USA, E-mail: hxl15@psu.edu
Citation: Lu H, Xie Z, Liu J, Lin L. Studies on Multiplex RT-PCR for Detection of Avian Influenza Virus Type A Group and Specific H5 and H7 Subtypes. J Veter Sci Med. 2013;1(2): 5.
Copyright © 2013 Lu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Veterinary Science & Medicine | ISSN 2325-4645 | Volume: 2, Issue: 1Submission: 25 October 2013 | Accepted: 25 November 2013 | Published: 29 November 2013
Avian Influenza (AI) is a highly contagious viral disease and occurs naturally in birds. Many wild birds are thought to be natural hosts of Avian Influenza Viruses (AIV) without necessarily becoming ill. There are 16 hemagglutinin (H-H16) and 9 neuraminidase (N-N9) subtypes of AIV recognized to date [1,2]. Although AI can be caused by any AIV subtypes, the H5 and H7 subtypes are the most lethal strains and have caused most of AI outbreaks worldwide in domestic poultry and other avian species [3-5]. These two subtypes can mutate from low pathogenic strains to highly pathogenic strains and cause severe epidemic AI outbreaks in poultry [6-8]. A highly contagious and virulent strain known as H5N1 caused the initial H5N1 outbreak and human deaths in Hong Kong in 1997 [9-11]. The Highly Pathogenic Avian Influenza (HPAI) H5N1 virus has emerged and caused widespread outbreaks in many Asian countries since late 2003 and it remains genetically different from the 1997 isolates [12-14]. Although the H5N1 virus continues to cause sporadic human infections in H5N1 affected countries or regions, virus transmission from animals to humans has no evidence of community level spread [15]. The HPAI H5N1 outbreaks have been mostly in poultry and water fowls in Asian countries since 2003 and have spread to Europe, Middle East and Africa [4,5,16] after the Qinghai Lake H5N1 outbreak in wide birds in May 2005 [17]. The H5N1 virus is an avian origin virus but the ongoing epidemic and confirmed human deaths suggest that the H5N1 virus may eventually mutate to a human pathogen to cause a potential flu pandemic [10,12,15,18].
The TRizol extraction procedure was used for virus RNA extraction. Briefly, 1) Test samples (including AF, tissue homogenate, tracheal and cloacal swabs prepared in virus transport medium (VTM) were clarified by centrifugation at 1200 rpm at 4° C for 10 min; 2) A volume of 250 μl supernatant per sample was transferred to a 1.8 ml sterile centrifuge tube, and then 750 μl TRizol reagent (Life Technology, Gaithersburg, MD) was added to each tube and mixed thoroughly by vortex; 3) The virus-TRizol mixture was incubated at ambient temperature for 10 min, then 200 μl chloroform was added and mixed thoroughly by vortex; 4) The virus-TRizol-chloroform mixture was incubated at ambient temperature for 10 min and thereafter was centrifuged at 12000 rpm for 10 min; 5) The aqueous layer (containing RNA) of each sample was transferred to a new 1.8 ml centrifuge tube, then an equal volume of isopropanol was added to each sample tube and the tube was incubated at ambient temperature for 5-10 min (for RNA precipitation); 6) The tube was centrifuged at 12000 rpm for 15 min to obtain the precipitated RNA, the supernatant was discarded after centrifugation, and then 1000 μl of 75% ethanol was added to each tube for one time wash, then the tube was centrifuged at 12000 rpm for 15 min and then the supernatant was discarded; 7) The tube was uncapped to allow air dry in a biosafety cabinet for 10-20 min until a visible moisture was disappeared; 8) 25 μl RNase-free water (DEPC-treated distilled water) was added to each sample tube for RNA elusion, then the RNA was ready for RT-PCR assay or was stored at -20° C or -80° C until use.
H5 and H7 subtype-specific primers
Se and Sp of the mRT-PCR
Studies on Multiplex RT-PCR for Detection of Avian Influenza Virus Type A Group and Specific H5 and H7 Subtypes
Huaguang Lu1*, Zhiqin Xie2, Jiabo Liu2 and Lin Lin1
- 1Animal Diagnostic Laboratory, Department of Veterinary and Biomedical Science, Pennsylvania State University, USA
- 2Guangxi Veterinary Research Institute, China
*Address for Correspondence: Huaguang Lu, Animal Diagnostic Laboratory, Department of Veterinary and Biomedical Science, Pennsylvania State University, USA, E-mail: hxl15@psu.edu
Citation: Lu H, Xie Z, Liu J, Lin L. Studies on Multiplex RT-PCR for Detection of Avian Influenza Virus Type A Group and Specific H5 and H7 Subtypes. J Veter Sci Med. 2013;1(2): 5.
Copyright © 2013 Lu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Veterinary Science & Medicine | ISSN 2325-4645 | Volume: 2, Issue: 1Submission: 25 October 2013 | Accepted: 25 November 2013 | Published: 29 November 2013
Abstract
A multiplex Reverse Transcription Polymerase Chain Reaction (mRT-PCR) for Avian Influenza Virus (AIV) has been developed to simultaneously Detect Nucleoprotein (NP) genes common to all AIV subtypes or type A influenza viruses by AIV NP gene or group primers and hemagglutinin (HA) protein genes specific to H5 and H7 subtypes by H5 and H7 primers, respectively. The AIV group primer bases were selected from conserved regions of AIV NP genes determined by sequence comparison and alignment using DNA STAR primer software (DNA STAR Inc., WI). Primers specific to H5 and H7 subtypes were designed on conserved sequences of HA protein genes of H5 and H7 subtypes, respectively. The mRT-PCR products amplified by AIV group primers gave a 750bp fragment, by H7 primers gave a 518bp fragment and by H5 primers gave a 410bp fragment. Evaluation test of the mRTPCR for AIV indicated that all AIV subtypes H1 through H7 and H9 tested were specifically amplified by AIV group primers, H5 subtypes were amplified by both group primers and H5 primers, and H7 subtypes were amplified by both group and H7 primers. Non-H5 or H7 subtypes were not amplified by the use of H5 or H7 primers. Samples contained both H5 and H7 subtypes were amplified by all 3 primer sets of AIV group, H5 and H7 primers simultaneously in one assay. Other avian viruses had no reaction to any of the AIV primers. The mRT-PCR for AIV detected as low as 2 x 10-3 ng of RNA extracted from H7N2 and H5N2 subtypes or as low as 2.2-2.5 Log10 ELD50 of these two viruses. Sensitivity (Se) and Specificity (Sp) of the mRT-PCR for AIV were measured as 92% and 100%, respectively, in comparison with virus isolation in Embryonated Chicken Eggs (ECE).Keywords
Avian influenza virus; Hemagglutinin; Nucleoprotein; Multiplex RT-PCR; PrimersAbbreviations
AI: Avian Influenza; AIV: Avian Influenza Virus; ECE: Embryonated Chicken Eggs; HA: Hemagglutinin; HI: Hemagglutination Inhibition; mRT-PCR: multiplex Reverse Transcription Polymerase Chain Reaction; NP: Nucleoprotein; Se: Sensitivity; Sp: Specificity; SPF: Special-Pathogen-Free (SPF); VI: Virus IsolationAvian Influenza (AI) is a highly contagious viral disease and occurs naturally in birds. Many wild birds are thought to be natural hosts of Avian Influenza Viruses (AIV) without necessarily becoming ill. There are 16 hemagglutinin (H-H16) and 9 neuraminidase (N-N9) subtypes of AIV recognized to date [1,2]. Although AI can be caused by any AIV subtypes, the H5 and H7 subtypes are the most lethal strains and have caused most of AI outbreaks worldwide in domestic poultry and other avian species [3-5]. These two subtypes can mutate from low pathogenic strains to highly pathogenic strains and cause severe epidemic AI outbreaks in poultry [6-8]. A highly contagious and virulent strain known as H5N1 caused the initial H5N1 outbreak and human deaths in Hong Kong in 1997 [9-11]. The Highly Pathogenic Avian Influenza (HPAI) H5N1 virus has emerged and caused widespread outbreaks in many Asian countries since late 2003 and it remains genetically different from the 1997 isolates [12-14]. Although the H5N1 virus continues to cause sporadic human infections in H5N1 affected countries or regions, virus transmission from animals to humans has no evidence of community level spread [15]. The HPAI H5N1 outbreaks have been mostly in poultry and water fowls in Asian countries since 2003 and have spread to Europe, Middle East and Africa [4,5,16] after the Qinghai Lake H5N1 outbreak in wide birds in May 2005 [17]. The H5N1 virus is an avian origin virus but the ongoing epidemic and confirmed human deaths suggest that the H5N1 virus may eventually mutate to a human pathogen to cause a potential flu pandemic [10,12,15,18].
AIV surveillance and rapid diagnosis play an essential role on AIV control strategies. While the historic and conventional laboratory methods for AIV isolation and identification are continuously used worldwide as standard assays, emerging novel technologies are rapidly being applied to support AIV surveillance and diagnosis [19]. During the past decade or recent years, many new AIV diagnostic tests including monoclonal antibody-based antigen capture assays [20-22] and new molecular assays such as real-time reverse transcription polymerase chain reaction (rtRT-PCR) [23,24], realtime reverse transcription loop-mediated isothermal amplification (rtRT-LAMP) [25], duplex real-time reverse transcriptase PCR (drtRT-PCR) [26], and rapid PCR-based molecular pathotyping of AIV H5 and H7 subtypes [27] have been developed to meet the urgent need of rapid and accurate diagnosis during outbreaks. This paper describes our recent studies in the development of a multiplex RT-PCR (mRT-PCR) for detecting nucleoprotein (NP) genes general to all AIV subtypes and hemagglutinin (HA) protein genes specific to H7 and H5 subtypes simultaneously in one assay.
Materials and Methods
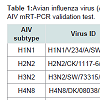
VirusesReference strains and field isolates of AIV including H1N1, H2N2, H3N2, H4N8, H5N2, H5N3, H5N9, H6N8, H7N2 and H9N2 subtypes stored at Wiley laboratory at the Pennsylvania State University were used for this study (Table 1). Each AIV strain was propagated in 9-11 day old Special-Pathogen-Free (SPF) Embryonated Chicken Eggs (ECE) following standard procedure [2,28]. Allantoic Fluid (AF) was harvested 72 hrs post inoculation and tested for the presence of AIV by the hemagglitinin (HA) test, Indirect Fluorescent Antibody (IFA) test and Dot-ELISA using AIV group-, H7 and H5 subtype-specific monoclonal antibodies [20,21,29,30].
Primer design
The DNA STAR primer software (DNA STAR Inc.,WI 53715) was used to design AIV group primers general to all subtypes of AIV and subtype-specific primers to H7 and H5 subtypes only. Each pair of primer bases were selected by comparison and alignment of sequences retrieved from GeneBank of the National Center Biotechnology Information (NCBI).
RT-PCR
Reverse Transcription (RT) master mixture was prepared using GeneAmp Gold RNA PCR Core Kit (Cat log # 4312765, Applied Biosystems, Foster city, CA). One reaction of RT master mixture contained 2 μl of 25 mM MgCl2, 4 μl of 5x PCR buffer, 2 μl of 10 mM deoxynucleoside triphosphate (dNTP), 2 μl of 100 mM DTT, 1 μl (20 units) of RNase inhibitor, 1 μl (0.5 μM) Random hexamers, 1 μl (50 units) of Moloney Murine Leukemia Virus (MuLV) reverse transcriptase and 50 ng of RNA, and 5 μl of sterile DEPC-treated water. A total volume of 18 μl RT mater mixture and 2 μl of RNA sample were mixed thoroughly for one RT reaction. The RT thermal cycle was programmed for one cycle at 25° C for 10 min, 42° C for 60 min, 99° C for 5 min and 4° C for 5 min. After completion of the RT reaction, 6 μl of of 25 mM MgCl2, 8 μl of 10 x RT-PCR buffer, 2.5 units of Ampli Taq DNA Polymerase, 100 pmol of AIV group primers, 100 pmol of H7 primers, 100 pmol of H5 primers, and sterile DEPC-treated distilled water were added to each tube to make a total volume of 100 μl per tube reaction. The reaction mixture was denatured at 94° C for 5 min, then the PCR thermal cycles were programmed for 35 cycles at 94° C for 1 min, 55° C for 1 min and 72° C for 1 min. One extension cycle was set at 72° C for 10 min and finally maintained at 4° C.
Electrophoresis
Horizontal agarose gel electrophoresis was used to analyze amplified DNA or PCR products. A 1.0% to 1.2% molecular grade agarose gel was prepared in 1X tris-borate buffer (45 mM tris-bonate, 1 mM EDTA) and 0.5 μg/ml ethidum bromide was added during preparation. A 10 μl PCR product was mixed with 2 μl loading dye per sample for gel loading. Gel electrophoresis was conducted at 100- 120 V with 30 A of power supply. Results were visualized with UV light, photographed with GenCam device and analyzed with GenCam software (Spectronics Corporation, Westbury, NY).
Sensitivity and Specificity Evaluation of the mRT-PCR for AIV detection
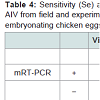
Sensitivity (Se) and Specificity (Sp) evaluation [31] of the mRTPCR for AIV detection were conducted by testing field specimens from 1998 and 2004 AIV positive cases and experimental samples from AIV-infected SPF chickens conducted in a previous study [32]. These samples had been saved at -80° C freezer in our laboratory. A standard reference test of Virus Isolation (VI) in ECE was used in comparison with the mRT-PCR and calculation of its Se and Sp [16]. Furthermore, serial dilutions of H5N2 (H5N2/CK/PA/85) and H7N2 (H7N2/CK/3779-2/PA/97) virus titers in mean embryo lethal dose (ELD50) and extracted virus RNAs were made for testing a minimum detectable amount of the virus titers or virus RNA by the mRT-PCR. Additionally, the mRT-PCR was evaluated by testing other avian viruses to certain that it would not react to other avian viruses including Infectious Bronchitis Virus (IBV), Infectious Laryngotrachitis Virus (ILTV), Fowl Adenovirus Type 1 (FAV-1), Paramyxovirus Type 1 (PMV-1), avian reovirus and poxvirus.
Results
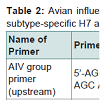
AIV group primersAIV group primer bases were selected by sequence comparison and alignment from 8 NP gene sequences of X52262 (H1N1),AY210102 (H2N2), AY210238 (H3N2), M63781 (H4N8), AF098628 (H5N2), AY221549 (H5N1), AY342427 (H7N7) and AY496851 (H9N2) stored at the GeneBank of NCBI. AIV group upstream primer consisted of 20 bases from 5’-AGC AGC ACA AAG AGC AAT GA-3’ and downstream primer consisted of 24 bases from 5’-ACT CAT GTC AAA GGA GGG CAC GAT-3’ (Table 2). The PCR product amplified by the AIV group primers is 750bp long.
Primers specific to H5 and H7 subtypes of AIV were designed on conserved sequences of H5 and H7 virus HA genes, respectively. Thirteen virus strains retrieved from GeneBank were used for alignment analysis of conserved regions of HA genes and designing specific primers to H5 and H7 subtypes. GeneBank numbers of the 13 virus trains including 5 different H5 subtypes for H5 primers,5 different H7 subtypes for H7 primers and 3 other subtypes for comparison are DQ497678 (H5N1), AY497086 (H5N2), AY296085(H5 N3), AY296086 (H5N8), AF194992 (H5N9), CY005983 (H7N1), AY240921 (H7N2), AY240894 (H7N3), CY005973 (H7N5), AB243420 (H7N7), CY010844 (H1N1), CY011688 (H3N2), DQ681216 (H9N2). H5 subtype-specific upstream primer consisted of 24 bases from 5’-TCA ATC CCA GAA ATA GCC ACC AGA-3’ and downstream primer consisted of 23 bases from 5’-ATC CAC CAT TCC TTG CCA TCC TC-3’. H7 subtype-specific upstream primer consisted of 24 bases from 5’-CGA TAG TAA GTT CCC TGC CAT TCC-3’ and downstream primer consisted of 24 bases from 5’-CAT TCT CCC TTA GTT GTT TCC TGA-3’. PCR products amplified by H5 and H7 primers were designed to amplify products of 410bp and 518bp, respectively (Table 2).
mRT-PCR for AIV
Ten subtypes of AIV reference strains and field isolates including H1N1, H2N2, H3N2, H4N8, H5N2, H5N3, H5N9, H6N8, H7N2and H9N2 (Table 1) were each tested by the mRT-PCR using AIV group, H7 and H5 multi-primer sets. All 10 subtypes were amplified by AIV group primers giving a product of 750bp in size, H7 subtype was also amplified by H7 primers giving a product of 518bp, and H5 subtype was also amplified by H5 primers giving a product of 410bp. A sample containing H7 and H5 subtypes was amplified by each of the 3 multi-primer sets simultaneously. Non-H7 and H5 subtypes were not amplified with the H7 and H5 primers (Figure 1).
Figure 1: AIV mRT-PCR using multiplex primer sets of AIV group, H7 and H5 subtype-specific primers for AIV subtypes H1N1 (1), H2N2 (2), H3N2 (3), H4N8 (4), H5N2 (5), H5N3 and H7N2 mix (6), H5N9 (7), H6N8 (8), H7N2 (9), H7N2 (10) and H9N2 (11). Mk = marker, 750bp = AIV group, 518bp = H7 subtype, 410bp = H5 subtype.
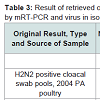
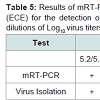
Three original H2N2 positive cloacal swabs collected from Pennsylvania (PA) poultry in 2004 and 8 H7N2 positive tissue specimens including tracheas, cecal tonsils and oviducts collected from 3 case submissions (A,B,C) during the 1998 H7N2 outbreak in PA poultry, which had been stored at -80 ° C freezer in our laboratory, were processed for AIV detection by the mRT-PCR. Two of the 3 H2N2 positive cloacal swabs from 2004 were positive to AIV group primers only and 5 of the 8 H7N2 positive tissues from 1998 were positive to both AIV group primers and H7 primers (Figure 2). These samples were retested by VI in ECE. Results of VI and mRT-PCR agreed each other in testing these 11 samples (Table 3). Furthermore, 22 positive and 14 negative original tracheal, cloacal and environment swab pools retrieved from a previous study of H7N2 virus in SPF chickens [25] were retested by both mRT-PCR and VI assays. Of the 22 H7N2 positive swab pools, 17 were positive by mRT-PCR and 19 were positive by VI. The 14 negative samples remained negative by both assays (Table 3). In comparison with VI results in testing the total number of 47 field and experimental samples, the mRT-PCR achieved 92% of Se and 100% Sp (Table 4). Additionally, three H5 subtypes of H5N2, H5N3 and H5N9 were successfully detected by both group primers and H5 primers of the mRT-PCR, 7 field isolates of H7N7 subtype were detect successfully detected by both group primers and H7 primers, and over 200 virus isolation negative AIV surveillance samples were all tested negative by the mRT-PCR. Furthermore, the mRT-PCR detected as low as 2.2 Log10 ELD50 of H7N2 virus (H7N2/ CK/3779-2/PA/97) and 2.5 Log10 ELD50 of H5N2 virus (H5/N2/CK/ PA/85), which were 1 Log10 less sensitive than that by VI (Table 5). Ten-fold serial dilutions were made on virus RNAs extracted from a mixture of H5N2 and H7N2 viruses (1:1) and a minimum amount of 2 x 10-3 ng RNAs was detected by the mRT-PCR (Figure 3). The mRT-PCR for AIV was specific to AIV as it did not amplify any other avian viruses tested including PMV-1, IBV Mass and Conn serotypes, ILTV, fowl adenovirus, avian reovirus and poxvirus.
Figure 2: AIV mRT-PCR for the detection of AIV from retrieved field samples of H2N2 and H7N2 positive cases. Samples 1, 2 and 3 were original H2N2 positive cloaca swabs collected from PA poultry in 2004; samples 4 (trachea), 5 (lung) and 6 (oviduct) were original tissues of case-A (H7N2 positive) from the 1998 H7N2 outbreak in PA; samples 7 (trachea), 8 (lung) and 9 (oviduct) were original tissues of case-B (H7N2 positive); and samples 10 (trachea/ lung) and 11 (oviduct) were original tissues of case-C (H7N2 positive). Mk = marker, 750bp = AIV group, 518bp = H7 subtype.
Figure 3: Sensitivity test of mRT-PCR for detecting 10-fold serial dilutions of virus RNAs extracted from H7N2 and H5N2 whole virus mix (1:1); 2x100ng(sample 4, positive), 2x10-1ng (sample 5, positive), 2x10-2ng (samples 6, positive), 2x10-3ng (sample 7, positive) 2x10-4ng (sample 8, negative), 2x10-5ng ( sample 9, negative), 2x10-6ng (sample 10, negative). Mk = marker, sample 1 = 10ng RNA of H2N2 virus for AIV group primer control (750bp), sample 2 = 10ng RNA of H7N2 virus for H7 primer control (518bp), sample 3 = 10ng RNA of H5N2 virus for H5 primer control (410bp).
Discussion
Standard RT-PCR and rtRT-PCR assays by single primer pairs have been reported to detect influenza type A viruses or specific subtypes of AIV [23,24,27]. In this study, a mRT-PCR assay for AIV has been developed to detect general NP gens to all AIV subtypes and specific HA protein genes to H5 and H7 subtypes simultaneously in one assay using AIV multiple primer pairs, which provides a rapid diagnosis for AIV and a confirmation of H5 or H7 subtypes at the same time. The multiplex primer sets for detecting AIV group and H5 and H7 subtypes showed that they had no interference to each other and they were as sensitive as a single primer pair for AIV detection. Therefore, the AIV group, H5 and H7 primers designed in this study can be used for multiplex primer sets and also a single primer set for AIV detection. This mRT-PCR for AIV achieved 92% Se and 100% in comparison with VI (Table 4) by testing a limited number of available old AIV samples in this study. Further studies to test a large number of samples from fresh AIV filed cases or experimental birds will provide more valuable data for its Se and Sp evaluation. Less positive results in retesting the saved or old AIV positive samples by VI andmRT-PCR in this study (Table 3) suggest that storage of original AIV positive samples over a period of time may cause decreasing virus titers to become undetectable, particularly those containing low virus titers. The old AIV positive samples may limit evaluation studies or accuracy of the mRT-PCR for AIV detection.AIV group, H5 and H7 primers developed in this study were evaluated on AIV reference strains and field isolates H1 through H7 and H9 subtypes available in our laboratory. Other subtypes including H8 and H10 through H16 were not tested and further evaluation on these subtypes should be conducted when they become available. Additionally, the H7 primer set was tested its specificity on H7N2 isolates only; it remains unknown that how the H7 primer set can detect other common H7 subtypes including H7N1, H7N3, H7N7 and H7N9. Thus different H7 subtypes shall be obtained for further evaluation and validation studies of the mRT-PCR for detection of all H7 subtypes. Nonetheless, the H5 primers were able to detect H5N2, H5N3, and H5N9 subtypes tested, and the H7 primers were able to detect the 7 field isolates of H7N2 subtype obtained during the 1998 and 2001 outbreaks in Pennsylvania poultry.
rtRT-PCR provides rapid test for AIV detection [23,24], but it requires expensive equipment and reagents. This mRT-PCR for AIV uses standard PCR equipment and reagents to achieve rapid detection of AIV, and it was nearly as sensitive as VI in chicken embryos (Table 3 & Table 5).
Acknowledgements
This research project was funded by Pennsylvania Department of Agriculture (PDA), Animal Health and Diagnostic Commission (AHDC) research program, PDA contract ME No. 443284.References
- Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, et al. (2005) Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79: 2814-2822.
- Swayne ED, Halvorson DA (2003) Influenza. In: Diseases of Poultry. (11th edn), Saif YM, Barnes HJ, Glisson JR, Fadly AM, McDougald LR, Swayne DE, Iowa State Press, Ames.
- Buisch WW, Hall AE, McDaniel HA (1984) 1983-1984 lethal avian influenza outbreak. In: Proc. 88th Annual Meeting of the United States Animal Health Association.
- FAO AIDEnews (2012) Update on the Avian Influenza situation. Issue No. 83.
- OIE (2013) Update Avian Influenza in Animals (Type H5). Updated: Oct 25.
- Banks J, Speidel ES, Moore E, Plowright L, Piccirillo A, et al. (2001) Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch Virol 146: 963-973.
- Ohuchi M, Orlich M, Ohuchi R, Simpson BEJ, Garten W, et al. (1989) Mutations at the cleavage site of the hemagglutinin alter the pathogenicity of influenza virus a/chick/penn/83 (H5N2). Virology 168: 274-280.
- Horimoto T, Rivera E, Pearson J, Senne D, Krauss S, et al. (1995) Origin and Molecular Changes Associated with Emergence of a Highly Pathogenic H5N2 Influenza Virus in Mexico. Virol 213: 223-230.
- Banks J, Speidel E, Alexander DJ (1998) Characterization of an avian influenza A virus isolated from a human is an intermediate host necessary for the emergence of pandemic influenza viruses. Arch Virol 143: 781-787.
- Class EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, et al. (1998) Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351: 472-477.
- Subbarao K, Klimov A, Katz J, Regnery H, Lim W, et al. (1998) Characterization of an avian influenza A ( H5N1) virus isolated from a child with a fatal respiratory illness. Science 279: 393-396.
- Campitelli L, Ciccozzi M, Salemi M, Taglia F, Boros S, et al. (2006) Influenza virus evolution: a comparison of different epidemics in birds and humans (1997-2004). J Gen Virol 87: 955-960.
- Cheung CL, Rayner JM, Smith GJ, Wang P, Naipospos TS, et al. (2006) Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. J Infect Dis 193: 1626-1629.
- Mase M, Eto M, Tanimura N, Imai K, Tsukamoto K, et al. (2005) Isolation of a genotypically unique H5N1 influenza virus from duck meat imported into Japan from China. Virology 339: 101-109.
- WHO (2011) WHO Comment on the importance of global monitoring of variant influenza viruses.
- Webster RG, Guan Y, Poon L, Krauss S, Webby R, et al. (2005) The spread of the H5N1 bird flu epidemic in Asia in 2004. Arch Virol Suppl 19: 117-129.
- Zhou JY, Shen HG, Chen HX, Tong GZ, Liao M, et al. (2006) Characterization of a highly pathogenic H5N1 influenza virus derived from bar-headed geese in China. J Gen Virol 87: 1823-1833.
- Webster RG, Shortridge KF, Kawaoka Y (1997) Influenza: interspecies transmission and emergence of new pandemics. FEMS Immunol Med Microbiol 18: 275-279.
- Charlton B, Crossley B, Hietala S (2009) Conventional and future diagnostics for avian influenza. Comp Immunol Microbiol Infect Dis 32: 341-350.
- Lu H (2003) A longitudinal study of a novel Dot-ELISA for detection of avian influenza virus. Avian Dis 47: 361-369.
- Lu H, Lin L, Wang R, Li Y, Scheuchenzuber B, et al. (2012) Development of H5 Subtype-specific Monoclonal Antibodies (MAb) and Mab-based Assays for Rapid Detection of H5 Avian Influenza. Health 4: 923-926.
- Woolcock PR, Cardona CJ (2005) Commercial immunoassay kits for the detection of influenza virus type A: evaluation of their use with poultry. Avian Dis 49: 477-481.
- Spackman E, Senne DA, Bulaga LL, Myers T J, Perdue ML, et al. (2003) Development of real-time RT-PCR for the detection of avian influenza virus. Avian Dis 47: 1079-1082.
- Sidoti F, Rizzo F, Costa C, Astegiano S, Curtoni A, et al. (2010) Development of Real Time RT-PCR Assays for Detection of Type A Influenza Virus and for Subtyping of Avian H5 and H7 Hemagglutinin Subtypes. Mol Biotechnol 44: 41-50.
- Postel A, Letzel T, Frischmann S, Grund C, Beer M, et al. (2010) Evaluation of Two Commercial Loop-Mediated Isothermal Amplification Assays for Detection of Avian Influenza H5 and H7 Hemagglutinin Genes. J Vet Diagn Invest 22: 61-66.
- Trevino C, Bihon S, Pinsky BA (2011) A Synonymous Change in the Influenza A Virus Neuraminidase Gene Interferes with PCR-Based Subtyping and Oseltamivir Resistance Mutation Detection. J Clin Microbiol 49: 3101-3102.
- Leijon M, Ullman K, Thyselius S, Siamak Zohari S, Pedersen JC, et al. (2011) Rapid PCR-Based Molecular Pathotyping of H5 and H7 Avian Influenza Viruses. J Clin Microbiol 49: 3860-3873.
- Centers for disease Control (1982) Concepts and Procedures for Laboratory-based Influenza Surveillance. Center for Disaese Control, Department of Health and Human Services, Washington, DC.
- Thayer SG, Beard CW (1998) Serological procedures. In: A Laboratory Manual for the Identification of Avian Pathogens. (4th edn), published by The American Association of Avian Pathologists.
- Wang X, Castro AE, Castro MD, Lu H, Weinstock D, et al. (2000) Production and evaluation criteria of specific monoclonal antibodies to the hemagglutinin of the H7N2 subtype of avian influenza virus. J Vet Diagn Invest 12: 503-509.
- Schwabe CW, Riemann H, Franti CE (1977) Epidemiology in Veterinary Practice. In Chapter 5 of Mathematical Approach: Test sensitivity and specificity, Philadelphia: Lea & Febiger.
- Lu H, Castro AE (2004) Evaluation of the infectivity, length of infection and immune response of a low-pathogenicity H7N2 avian influenza virus in SPF Chickens. Avian Dis 48: 263-270.