Journal of Pharmaceutics & Pharmacology
Download PDF
Research Article
Lysosomotropic Action of Amantadine: Basis for Treatment of COVID-19
Smieszek SP*, Przychodzen BP and Polymeropoulos MH
Vanda Pharmaceuticals Inc., Washington, DC, USA
*Address for Correspondence:
Smieszek SP, Vanda Pharmaceuticals Inc., 2200 Pennsylvania NW, Suite 300-
E, Washington, DC 20037, USA; Email: sandra.smieszek@vandapharma.com
Submission: 06 August 2020;
Accepted: 04 September 2020;
Published: 10 September 2020
Copyright: © 2020 Smieszek SP, et al. This is an open access article
distributed under the Creative Commons Attribution License, which
permits unrestricted use, distribution, and reproduction in any medium,
provided the original work is properly cited.
Abstract
SARS-coronavirus 2 is the causal agent of the COVID-19 outbreak. SARSCov-
2 entry into a cell is dependent upon binding of the viral Spike (S)
protein to cellular receptor and on cleavage of the spike protein by
the host cell proteases such as Cathepsin L and Cathepsin B (CTSL/B).
They are crucial elements of lysosomal pathway and both enzymes
are almost exclusively located in the lysosomes. CTSL disruption offers
potential for CoVID-19 therapies. The mechanisms of disruption include:
decreasing expression of CTSL, direct inhibition of CTSL activity and
modification of the CTSL environment (increase pH in the lysosome).
We have conducted a high throughput drug screen gene expression
analysis to identify compounds with the capacity to downregulate the
expression of CTSL/CTSB. One of the most significant results shown to
downregulate the expression of the CTSL gene is Amantadine(10uM).
We confirmed Amantadine’s lysosmal trapping capacity in an invitro
Lysosomal Trapping Assay. In addition, to downregulating CTSL,
Amantadine disrupts the lysosomal pathways, hence, interferes with
the capacity of the virus to replicate. It acts as a lysosomotropic agent
altering the CTSL functional environment. We propose that Amantadine
could decrease the viral load in SARS-CoV-2 positive patients and as
such it may serve as a potent therapeutic decreasing the replication
and infectivity of the virus likely leading to better clinical outcomes.
Clinical studies are currently needed to examine the therapeutic
efficacy of Amantadine in COVID-19 infection.
Introduction
Recently a novel type of highly virulent beta-coronavirus was
discovered in patients with pneumonia of unknown cause. Severe
Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) as detected
by sequencing of the samples was found to be the cause of a severe
respiratory disease in humans [1]. The outbreak of COVID-19
resulted in a global epidemic with the number of confirmed cased
surpassing 722, 000 in March 2020.
The SARS-CoV-2 genome shares about 80% similarities with
SARS-CoV and is even more similar (96%) to the bat coronavirus
BatCoVRaTG13 [2]. Corona viruses are characterized by large genetic
diversity and frequent recombination of their genomes and hence
pose a challenge in terms of public health, currently based on 1455
viral genomes and predicted 24.5 genetic substitutions per year [3].
Similar to SARS-CoV, SARS-Cov-2 enters the cell by the
means of binding to cellular receptor(s) including the Angiotensin-
Converting Enzyme 2 (ACE2) membrane bound protein [4]. Host
protease dependence of SARS-CoV-2 entry is a critical step. SARSCoV
takes advantage of the endosomal cysteine proteases CTSL and
CTSB [5,6]. CTSL is a peptidase that preferentially cleaves peptide
bonds with aromatic residues in P2 and hydrophobic residues in the
P3 position [7]. CTSL is active at pH 3-6.5, in the presence of thiol
and its enzymatic stability is dependent on ionic strength [7]. CTSL
proteolysis is a crucial mechanism for Ebola as well as SARS-CoV
for processing of viral glycoprotein before cell membrane fusion [6].
Specifically, during cell membrane fusion, the S protein is cleaved by host cell proteases, exposing a fusion peptide of the S2 domain. This
leads to the fusion of viral and cellular membranes and the release of
the viral genome into the cytoplasm of the host cell.
Cleavage at both sites is believed to be necessary for viral entry
by endocytosis into the host cell. The S1/S2 cleavage site of SARSCoV-
2 is between the threonine and methionine at positions 696 and
697. This S1/S2 cleavage site is identical to that of SARS-CoV which
has been shown to be cleaved by CTSL, a lysosomal cysteine protease
encoded by the CTSL1 gene. SARS-CoV-2 also has a furin-like
protease cleavage site not found in SARS-CoV, between the arginine
and serine at positions 685 and 686. This site may be cleaved by furin
during viral egress. Interfering with the spike protein processing by
the host cell, whether by affecting the environment or modulating
gene expression levels, hence offers a potential therapeutic strategy.
Genetic variants within CTSL gene could in theory affect the
propagation capacity of the virus. Furthermore CTSL polymorphisms
could affect the susceptibility to SARS-CoV-2 where for example
individuals with certain genetic variant have reduced expression
of CTSL and in turn could be protected or have lower viral titers.
Additionally, elements of hosts Major Histocompatibility Complex I
(MHC I) and cytotoxic T Cell Lymphocytes (CTL)-mediated immune
responses might affect viral proliferation [8]. There are susceptibility
factors ranging from ethnicity background to age related groups, to
comorbid conditions [9,10].
In a report of results of an earlier segment of our investigations we
tested compounds that could help identify potential therapeutic agents
with the capacity to decrease expression or inhibit the expression of
the CTSL gene [11]. We identified Amantadine among top of the list
of significant compounds. We now further confirmed Amantadine’s
lysosmal trapping capacity in an in-vitro Lysosomal Trapping Assay.
Together these results provide a large body of evidence suggesting
potential efficacy of Amantadine in treatment of COVID-19.
Materials and Methods
Cell culture and drug treatment:
Drugs screening was carried out, the same one as applied in our previous study [12]. The retinal pigment epithelia cell line, ARPE-19/
HPV-16, was chosen to establish a database of drug profiles because
of its non-cancerous, human origin, with a normal karyotype. It can
also be easily grown as monolayer in 96-well plates. Compounds were
obtained from Sigma (St. Louis, MO) or Vanda Pharmaceuticals
(Washington, DC). Cells were aliquoted on 96-well plates (~2×10e5
cells/well) and incubated for 24 h prior to providing fresh media
with drug, or the drug vehicle (water, dimethyl sulfoxide, ethanol,
methanol, or phosphate-buffered saline solution). Drugs were diluted
1000 fold in buffered in Dulbecco’s Modified Eagle Medium: Nutrient
Mixture F-12 (D-MEM/F-12) culture medium (Invitrogen, Carlsbad,
CA) containing nonessential amino acids and 110 mg/L sodium
pyruvate. In these conditions, no significant changes of pH were
expected, which was confirmed by the monitoring of the pH indicator
present in the medium. A final 10 μM drug concentration was chosen
because it is believed to fit in the range of physiological conditions
[12]. Microscopic inspection of each well was conducted at the end of
the treatment to discard any samples where cells had morphological
changes consistent with apoptosis. We also verified that the drug had
not precipitated in the culture medium.Gene expression:
Cells were harvested 24 h after treatment and RNA was extracted
using the RNeasy 96 protocol (Qiagen, Valencia, CA). Gene
expression for 22,238 probe sets of 12,490 genes was generated with
U133A2.0 microarrays following the manufacturer’s instructions
(Affymetrix, Santa Clara, CA). Drugs were profiled in duplicate or
triplicate, with multiple vehicle controls on each plate. A total of 708
microarrays were analyzed including 74 for the 18 antipsychotics, 499
for the other 448 compounds, and 135 for vehicle controls. The raw
scan data were first converted to average difference values using MAS
5.0 (Affymetrix). The average difference values of both treatment
and control data were set to a minimum of 50 or lower. For each
treatment category, all probe sets were then ranked based on their
amplitude or level of expression relative to the vehicle control (or
the average of controls was selected when more than one was used).
Amplitude was defined as the ratio of expression (t−v) / [(t+v) / 2]
where t corresponds to treatment instance and v to vehicle instance.In vitro hepatocyte lysosomal trapping studies:
This protocol was designed to evaluate Amantadine for lysosomal
trapping potential in immortalized hepatocytes (Fa2N-4 cells). Fa2N-4 cells are immortalized human hepatocytes that retain expression
and function of lysosomes and can be used to evaluate accumulation
of compounds in lysosomes. Specifically, the test article was incubated
with Fa2N-4 cells in the presence or absence of ammonium chloride
(an inhibitor of lysosomal trapping). The amount of test article that
accumulates in the cells was quantified by LC-MS/MS. Incubations
of propranolol (a known lysosomotropic drug) with and without
ammonium chloride were used as positive controls.Results
Drug screening:
With the aim of discovering potential pharmaceutical agents
capable of affecting transcriptional expression levels of CTSL
implicated in SARS-CoV and SARS-CoV2 Pathophysiology, we
screened 466 compounds belonging to 14 different therapeutic classes.
Screening was conducted using human retinal pigment epithelia cell
line (ARPE-19) and gene expression changes were collected across
12,490 genes. The ARPE-19 cell line was initially selected as a well
suited model for the study of compounds that affect neuronal type
cells, in particular antipsychotics. Here, we describe the discovery of a
CTSL/B, lysosomotropic signature which might give insights into the
therapeutic potential of the tested compounds.We analyzed the expression profiles of CTSL across all 466
compounds tested. In order to find positive hits we selected only
those results that showed a reduction of expression of CTSL (1.5 -fold
difference).There were no drugs that decreased CTSL expression by
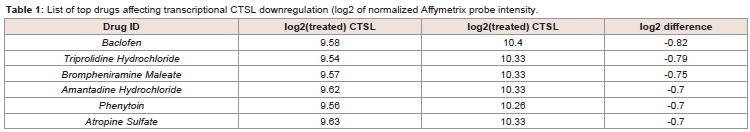
more than 40%. Between the most 5 potent compounds were drugs
from various therapeutic areas - muscle relaxant, antihistamine, antiepileptic,
anticholinergic and antiviral (Table 1). Top results (top
5 of 466) included Amantadine hydrochloride, an established and
safe antiviral agent that was previously used to treat patients with
influenza A.
Table 1: List of top drugs affecting transcriptional CTSL downregulation (log2 of normalized Affymetrix probe intensity.
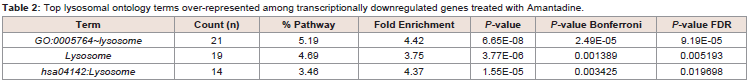
Table 2: Top lysosomal ontology terms over-represented among transcriptionally downregulated genes treated with Amantadine.
Due to its high lipophilicity, Amantadine can cross lysosomal
membranes and accumulate in lysosomes acting at higher
micromolar concentrations as lysosomotropic alkalinizing agent [9-13]. Amantadine inhibits influenza A replication at low micromolar
concentration, by blocking M2 ion channel protein which acidifies
the virus interior and releases its nucleoprotein [9-14]. It causes pH
alteration which ultimately abrogates membrane fusion a necessary
step for virus replication [13,14]. Other lysosomotropic drugs affect
lysosomes through lysosome membrane permeabilization and accumulation also blocking of Ca2+ signaling, and enzyme activity
inhibition orstorage material accumulation [15]. Since Amantadine
behaves as a lysosomotropic substance that passes easily through the
lysosome membrane of SARS-CoV-2 virus and accumulates, where
it could increase the pH of lysosome and thus inhibit the protease
activities [15]. Moreover Amantadine may directly affect viral entry
by down-modulating CTSL and other lysosomal pathway genes. The
PK profile of the drug makes it particularly suitable for administration
to humans. Plasma concentration is in the range of 200-800 ng/mL
depending on the formulation and dosing regimen. Plasma half-life
is 17 h (range: 10-25 h) with renal clearance as main elimination
mechanism. Amantadine HCl [immediate release] is available as a
100-mg tablet and 50 mg/5 mL syrup and is typically administered
twice daily [16]. Human cells in tissue culture readily tolerated
Amantadine up to a concentration of 100 ug/mL (~657 uM).
Since CTSL was not the top differentially-expressed transcript, we
decided to extend our analysis to all the genes that were downregulated
by Amantadine. Among the top 500 differentially expressed probes
(383 genes, all with at least 50% expression reduction) we have found
21 genes related to lysosomal terms using David Enrichment tool
(GO:005764, p=2.49x10-5). Moreover, the top significant pathway by
ENRICHR enrichment analysis toolwasthe KEGG lysosome pathway.
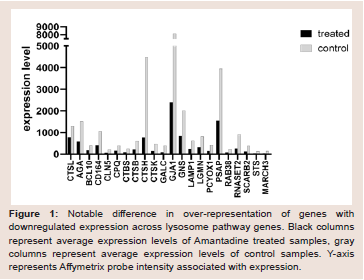
Amantadine’s significant effect upon lysosome pathway genes is
shown on Figure 1 and Table 2. Figure 1 displays notable difference in
over-representation of genes with downregulated expression across
lysosome pathway genes. Table 2 displays top lysosomal ontology
terms over-represented among transcriptionally downregulated
genes treated with Amantadine.
Figure 1: Notable difference in over-representation of genes with
downregulated expression across lysosome pathway genes. Black columns
represent average expression levels of Amantadine treated samples, gray
columns represent average expression levels of control samples. Y-axis
represents Affymetrix probe intensity associated with expression.
Lysosomal trapping assay:
Lipophilic and amphiphilic drugs with ionizable amines can
accumulate in lysosomes - a process known as lysosomal trapping
[17]. To test the capacity of Amantadine to act as lysosomotropic
agent we have conducted an in-vitro hepatocytes lysosomal trapping
assay as described previously in literature [17,18]. We focused on the
difference in uptake and hence the amount of test compound trapped
in lysosomes. We confirmed that Amantadine showed lysosomal
uptake and lysosomal trapping capacity. Circa 50% of Amantadine
was trapped in lysosomes at 1 μM and showed significant saturation at higher concentrations. Specifically, the uptake of Amantadine
(1, 10 and 100 μM) in Fa2N-4 cells was concentration dependent
and was reduced up to 46.7 and 40.5% at 1 μM in the presence of
ammonium chloride at the 10 and 30 min incubation period,
respectively (Supplemental Figure 1). The uptake was marginally
reduced at 10 and 100 μM. These results suggest the potential for
lysosomal trapping at low concentration. Hence, the experiments
confirmed Amantadine’s capacity to act as a lysosomotropic agent.Discussion
Decreasing the expression of CTSL is likely a potential mechanism
that would lower the capacity of the virus to enter the next host
cell. Another symbiotic, therapeutic mechanism is lysosomal pH
modulation that would further interfere with proteolytic spike
protein activation. Therapeutic agents capable of perturbing the
lysosomes, their function or microenvironment may offer protection
from the virus or decrease the severity of the symptoms. Given
that Amantadine not only down-regulates CTSL expression, but
a number of key lysosomal enzymes, we can now hypothesize that
lysosomal dysfunction induced by Amantadine administration could
be protective against viral entry and ultimately replication. Our
hypothesis is that people with certain lysosomal storage diseases may
be resistant to one of these viruses. Along these lines there is suggestive
evidence for this to be true. For example Niemann-Pick disease type
C1 lipid storage disorder offers resistance to Ebola in patient cell lines
[19,20]. Interestingly, bat species show selective sensitivity to Ebola
versus Marburg viruses [21].
Interfering with the lysosomal milieu can have protective effects
from coronavirus which we know uses CTSL, a pH sensitive enzyme,
to process the cleavage of the spike protein. Amantadine would be
predicted by physical and chemical properties to accumulate in the
lysosomes and raise pH, interfering with CTSL function. The gene
expression pattern reported in this paper suggests that a more general
lysosomal program is down-regulated by Amantadine, likely through
a common set of transcription factors. Additionally, Amantadine’s
property to accumulate in lysosomes, if effective, could reduce
viral load, decrease intra-host organ spread and decrease patientassociated
disease severity and progression. Importantly, the dose
of Amantadine that was tested in High Throughput Screen Assay is
within one order of magnitude of expected pharmacokinetic, clinical
profile (~5 uM). That would mean the drug can be administered per
existing, safe and approved label dosing.
Further studies including clinical trials are now required in order
to examine the role of Amantadine administration as a treatment for
COVID-19.
Acknowledgement
We thank all the reviewers for valuable comments and suggestions.