Journal of Nutrition & Health
Download PDF
Research Article
*Address for Correspondence: Yingying Le, Key Laboratory of Food Safety Research, Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China, Tel: +86-21-54920901; E-mail: yyle@sibs.ac.cn
Citation: Liu X, Qin S, Wang H, Le Y. Monitoring Genetically Modified Rice Using Multiplex PCR Coupled with Universal DNA Microarray. J Nutri Health. 2016;2(2): 6.
Copyright © 2016 Le et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Nutrition and Health | ISSN: 2469-4185 | Volume: 2, Issue: 2
Submission: 15 April, 2016 | Accepted: 30 June, 2016 | Published: 05 July, 2016
LDR reaction: 10 μl of multiplex PCR product was mixed with 1 μl Proteinase K (20 mg/ml), which was incubated at 70 °C for 10 min and quenched at 94 °C for 20 min. LDR was carried out in a final volume of 20 μl containing 10 μl Proteinase K digested multiplex PCR product, 20 mM Tris-HCl (pH 7.6), 25 mM potassium acetate, 10 mM magnesium acetate, 10 mM DTT, 1 mM NAD, 0.1% Triton X-100, 1 pmol each discrimination probe, 1 pmol each common probe, 0.5 μl 40 U/μl Taq DNA ligase (New England Biolabs, USA). The reaction was performed with 40 cycles at 94 °C for 30 s and 65 °C for 4 min.
Screening GM rice using multiple PCR-LDR-UA systemWe examined 9 rice samples from different producers randomly collected from different markets in Shanghai by using the multiple PCR-LDR-universal DNA array systems, no GM rice was detected data not shown).
Monitoring Genetically Modified Rice Using Multiplex PCR Coupled with Universal DNA Microarray
Xiaolei Liu1,2, Shengying Qin3, Hui Wang1,2 and Yingying Le1,2*
- 1Key Laboratory of Food Safety Research, Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China
- 2Key Laboratory of food Safety Risk Assessment, Ministry of Health, Beijing, China
- 3Bio-X Life Science Research Center, Shanghai Jiao Tong University, Shanghai, China
*Address for Correspondence: Yingying Le, Key Laboratory of Food Safety Research, Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China, Tel: +86-21-54920901; E-mail: yyle@sibs.ac.cn
Citation: Liu X, Qin S, Wang H, Le Y. Monitoring Genetically Modified Rice Using Multiplex PCR Coupled with Universal DNA Microarray. J Nutri Health. 2016;2(2): 6.
Copyright © 2016 Le et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Nutrition and Health | ISSN: 2469-4185 | Volume: 2, Issue: 2
Submission: 15 April, 2016 | Accepted: 30 June, 2016 | Published: 05 July, 2016
Abstract
We developed a multiplex polymerase chain reaction (PCR) coupled with ligation detection reaction and universal array system for screening genetically modified rice and further identifying specific transgenes in rice. Three common elements in transgenic rice (CaMV 35S promoter, FMV 35S promoter and nos terminator) with species and internal controls (rice actin, 18S rRNA) were used for screening transgenic rice from un-transgenic rice. Seven event-specific elements (Cpt1, Bt, Irp, Bar, OsVTE1, Xa21 and RCH10) were used for identifying specific transgenes in rice. This method was demonstrated to be effective and high throughput for the detection of transgenic rice. It can be applied not only for detecting commercial transgenic rice, but also for monitoring unauthorized transgenic rice.Keywords
Genetically modified organism; Transgenic rice; Multiplex PCR; Ligation detection reaction; MicroarrayAbbreviations
Bar: Bialaphos resistance gene; Bt: Cry1Ac gene from Bacillus thuringiensis; CaMV: Cauliflower Mosaic Virus; CptI: Cowpea trypsin Inhibitor gene; FMV: Figwort Mosaic Virus; GMO: Genetically Modified Organism; LDR: Ligation Detection Reaction; Lrp: Winged bean Lysine rich protein gene; nos: Agrobacterium tumefaciens nopaline synthase; OsVTE1: Rice tocopherol cyclase gene; PCR: Polymerase Chain Reaction; RCH10: Rice basic Chitinase gene; Xa21: Rice bacterial blight disease resistance geneIntroduction
Rice is cultivated in more than 100 countries and supplies staple food for nearly half of the world’s population, especially those living in developing countries. The intensively increasing world population has put a great burden on rice production. However, rice yield is severely influenced by biotic and abiotic factors, such as viruses, bacteria, pests, drought- and salt-stress. Compared with conventional breeding method, molecular method represents an increasingly important approach for genetic improvement of disease resistance, environmental adaptation, and nutrient content. Since the first development of transgenic rice in 1988 [1-3], rice has been engineered for enhanced resistance to disease, herbicides, environmental stress; and for improving nutritional quality [4,5]. Although other transgenic crops have been authorized for food and/or feed by many countries based on their own criteria for safety assessment, there are only seven authorized transgenic rice worldwide [http://www.isaaa.org/gmapprovaldatabase/]. China has been the largest producer and consumer of rice in the world. Genetically modified rice has a rapid development in recent years. However, no transgenic rice has been approved to be commercialized in China. Despite strict regulations, unauthorized transgenic rice has been occasionally released into the market. Consumers have been demanding appropriate information and labeling for foods derived from transgenic rice. Therefore, it’s urgent to develop methods for monitoring transgenic rice in market.To date, a wide variety of methods have been developed for detecting genetic modified (GM) crops. Techniques based on nucleic acid analysis have been widely applied for detecting GM crops. Conventional PCR and real-time PCR that amplify a single target have been used to qualitative and quantitative analysis of GMO, respectively [6,7]. Multiplex PCR employs several primer pairs in the same amplification reaction for a DNA sample and can simultaneously detect multiple target DNA sequences. It saves considerable time and effort in GM crop detection [8,9]. However, the identity of the PCR products of multiplex PCR need to be further verified by digestion of the DNA with restriction enzymes followed by fragment analysis, or DNA sequencing, capillary electrophoresis, or high-performance liquid chromatography. Oligonucleotide-based hybridization assay, also known as microarray or DNA chip, can simultaneously identify multiple nucleic acid sequences, thus offering the possibility for rapid and high throughput detection of GM crops. Different microarray approaches have been developed to be used in combination with multiplex PCRs to detect transgenes in different corps, food and feed [10,11].
PCR coupled with ligation detection reaction (LDR) and universal array (UA) has been proposed by Gerry et al. as a powerful tool for sequence discrimination. PCR-LDR-UA systems have been developed to detect low abundant DNA point mutation [12], DNA single nucleotide polymorphism [13], bacterial discrimination [13]. This approach allows for the unequivocal detection and amplification of several amplicons in a single experiment.
In the present study, we developed a multiplex PCR-LDR-UA system to detect GM rice. This system can determine both commercial and unauthorized transgenic rice and the identity of transgenes.
Materials and Methods
Rice seeds and DNA extractionSeeds of transgenic rice and their corresponding wild type rice were obtained from various laboratories in China (Supplementary Table 1). Genomic DNA was extracted with DNA extraction Kit for GMO Detection Ver.2.0 (Takara). The DNA concentration was measured by absorbance at 260 nm, and the quality of DNA was evaluated by the 260 to 280 nm absorption ratios. DNA samples with a ratio of 1.6-1.9 were used in the following assays. For PCR reactions, DNA concentration was adjusted to 50-100 ng/μl with double distilled water.
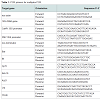
We designed two groups of PCR primers to amplify target genes with multiplex PCR system. The first group of primers was used for screening transgenic rice from un-transgenic rice. It consists of primers for amplification of common elements in transgenic rice (CaMV 35S promoter, FMV 35S promoter, nos terminator), rice species control and internal control (rice actin, 18S rRNA). The second group of primers was used for further identifying specific transgenes in rice. PCR primers were designed by MIT Whitehead Institute Primer 3.0 software. PCR primers met the following criteria: (1) have a highly approximation of melting temperature (Tm) between 55-60 °C; (2) amplification products span between 100-500 bp, and differ from each other for up to 50 bp ; (3) the homology of primers are as low as possible; (4) be compatible in the PCR mixture. The primers were synthesized by Invitrogen. Primer sequences are listed in Table 1.
Multiplex PCR reaction were carried out on the PTC-100 DNA engine (MJ Research, USA) in a total volume of 25 μl containing 50 ng of rice genomic DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3.0 mM MgCl2, 200 mM dNTP, 1 μM of each primer and 0.25 U Taq DNA polymerase. The cycling protocol consisted of denaturation at 95 °C for 5 min, followed by 40 cycles at 95 °C for 30 s, 55 °C for 40 s, 72 °C for 1 min, and a final extension at 72 °C for 10 min. PCR products were visualized by ethidium bromide staining in 1.5 % agarose gel. Each experiment was repeated at least three times.
LDR probe design, LDR and hybridization
We designed the discriminating probe and common probe specific to each amplicon of multiplex PCR as previously described (Table 2) [12,13]. The discriminating probe contains a Zip-code complement sequence (cZip) at the 5’-end which is complementary to a Zip Code sequence in the universal array. The common probe is phosphorylated at the 5’-termini and carries Cy3 fluorochromes at the 3-end). The Tm values of these probes were designed as close as possible, and the specificity of the probes was checked through BLAST program. During LDR, the two probes are hybridized adjacent to one another on the multiplex PCR-amplified target DNA and the nick between them is sealed by the ligase only if there is perfect complementary between the probes and target DNA sequences. The resulting products are hybridized onto the universal array slide where the cZip Code at the 5’end of the ligated oligonucleotide binds with the corresponding Zip code on the slide (Figure 1).
LDR reaction: 10 μl of multiplex PCR product was mixed with 1 μl Proteinase K (20 mg/ml), which was incubated at 70 °C for 10 min and quenched at 94 °C for 20 min. LDR was carried out in a final volume of 20 μl containing 10 μl Proteinase K digested multiplex PCR product, 20 mM Tris-HCl (pH 7.6), 25 mM potassium acetate, 10 mM magnesium acetate, 10 mM DTT, 1 mM NAD, 0.1% Triton X-100, 1 pmol each discrimination probe, 1 pmol each common probe, 0.5 μl 40 U/μl Taq DNA ligase (New England Biolabs, USA). The reaction was performed with 40 cycles at 94 °C for 30 s and 65 °C for 4 min.
The universal array slides with 5’-amino-modified Zip-codes were prepared as described previously [13]. The LDR reaction product was diluted to obtain 70 μl of hybridization mixture containing 5 × SSC and 0.1 mg/ml salmon sperm DNA, quenched at 95 °C for 5 min, chilled on ice immediately, and applied onto the universal array slide using Gene Frame slide chamber (ABgene, UK). Hybridization was carried out at 65 °C for 2 h. The slide was washed for 2 min in 1 × SSC, 0.1% SDS and then for 1 min in 0.05 × SSC by gently shaking at room temperature. Finally, the slide was spun at 1000 r.p.m. for 4 min. The image was scanning on a GMS418 scanner (Genetic Micro- Systems, USA).
Results
Design of PCR primers and establishment of multiplex PCR systemsMultiplex PCR requires that primers lead to amplification of unique regions of DNA, both in individual pairs and in combinations of many primers, under a single set of reaction conditions. We designed two groups of PCR primers to amplify target genes with multiplex PCR system. The first group of primers was used for screening transgenic rice from un-transgenic rice. It consists of primers for amplification of common elements in transgenic rice, rice species control and internal control. The second group of primers was used for further identifying specific transgenes in rice.
A large number of transgenic crop plants, such as soybean, maize and canola, contain the 35S promoter in the cauliflower mosaic virus (CaMV 35S) or figwort mosaic virus (FMV 35S), and Agrobacterium tumefaciens nopaline synthase (nos) terminator. These common elements have been also used in transgenic rice and cover more than 95% of GM rice. Therefore, we include primers targeting CaMV 35S, FMV 35S, nos terminator, rice actin and 18S rRNA in the first group for screening transgenic rice from un-transgenic rice. The specificity of each primer pair was tested in single PCR reaction with genomic DNA from transgenic rice and non-transgenic rice. Primer pairs for rice actin and 18S rRNA each amplified products of the expected lengths from the genomic DNA of both transgenic and nontransgenic rice. Each of primer pairs for CaMV 35S promoter, FMV 35S promoter, and nos terminator amplified products from target GM rice DNA, but not from non-transgenic rice DNA. All PCR products were confirmed by sequencing (data not shown). Then the first group of primers targeting rice actin, 18S rRNA, CaMV 35S promoter, FMV 35S promoter and nos terminator were included in one multiplex PCR reaction. By designing specific primers and adjusting reaction conditions, we simultaneously amplified all five targets of different lengths which could be detected by agarose gel electrophoresis, and the procedure was capable of distinguishing non-transgenic rice from transgenic rice (Figure 2A and data not shown). Primers targeting rice actin and 18S rRNA were included in the multiplex PCR reaction as controls for confirming the rice species, evaluating DNA quality and PCR efficacy, eliminating any possibility of false negatives, thereby increasing reliability.
The second group of primers targets specific transgenes in rice, including Bar (bialaphos resistance gene), Bt (cry1Ac gene from Bacillus thuringiensis), CptI (cowpea trypsin inhibitor gene), Xa21 (rice bacterial blight disease resistance gene), Rch10 (rice basic chitinase gene), Lrp (winged bean lysine rich protein gene), and OsVTE1 (rice tocopherol cyclase gene). The specificity of each primer pair was tested in single PCR reaction with genomic DNA from transgenic rice and non-transgenic rice. Each of primer pairs for Bar, Bt, CptI, Xa21, Rch10, Lrp, and OsVTE1 amplified products from target GM rice DNA, but not from non-transgenic rice DNA. All PCR products were confirmed by sequencing (data not shown). Then the seven pairs of primers in the second group were included in one multiplex PCR reaction to identify specific transgene(s) in rice. To develop an efficient multiplex PCR for detecting these transgenes simultaneously, multiple attempts were made to optimize reaction conditions, including adjusting the concentrations of the primers, the PCR buffer, magnesium chloride and deoxynucleotide, the cycling temperatures, and the amount of template DNA and Taq DNA polymerase. We successfully amplified 7 transgenic genes in a multiplex PCR according to their specific sequences (Figure 2B). Although multiplex PCR procedures have been developed for identification of some or most common transgenic lines of maize, soybean and canola, to our knowledge, this is the first study in which simultaneous amplification profiling has been described as a reliable tool for identifying transgenic lines of rice.
LDR and universal array
LDR is initially developed to recognize DNA point mutation or single nucleotide polymorphism using high-temperature enzyme, this procedure increase the specificity of the detection [12,13]. A ligation detection reaction universal array has been developed for detection of transgenic events in commercialized maize and soybean [15,16]. To verify the results of multiplex PCR, we established two groups of LDR-universal array systems. In the first system, the LDR probes targeted amplicons from common elements of transgenic rice (CaMV 35S promoter, FMV 35S promoter and nos terminator) and internal controls (rice actin and 18S rRNA). Each Zip sequence on the universal array slide has a complementary cZip sequence at the 5’end of the discriminating probe. The second system contains LDR probes targeting amplicons from specific transgenes in rice (Cpt1, Bt, Irp, Bar, osvte, Xa21 and RCH10), and Zip sequences on the universal array slide which has a complementary cZip sequence at the 5’end of the discriminating probe. Combination of LDR with universal array could validate the results of multiplex PCR to identify specific transgenes in rice. By using the first LDR-universal array system, we detected rice actin and 18S RNA in both nontransgenic and transgenic rice (Figures 3A and 3B), but detected FMV 35S promoter, CaMV 35S promoter and nos terminator only in transgenic rice (Figure 3B), validating the results of multiplex PCR and discriminating transgenic rice from nontransgenic rice. By using the second LDR-universal array system, we detected no transgenes in non-transgenic rice (Figure 4A), but detected Cpt1, Bt, Lrp, Bar, Osve, Xa21 and Rch10 in transgenic rice (Figure 4B), confirming the results of multiplex PCR and identifying the exogenous gene in transgenic rice.
Discussion
There are very few studies on screening of transgenic rice, especially unauthorized rice. In the present study, we developed multiplex PCR coupled with universal DNA microarray systems to screen and identify transgenic rice. Multiplex PCRs have been developed to detect GMOs coupled with DNA microarray analysis, which mainly focus on commercial GMOs, such as soybean, maize and canola [10,11,17]. These methods detect GMOs by using fluorescent dye- or biotin-labeled dNTP or PCR primers [11,17,18] in PCR system. Multiplex PCR can easily generate unspecific products, increasing the probability of having false positives and ambiguous results. The capture probes fixed on the chip are complementary to their specific targets. Different capture probes need to be designed according to the detecting targets. In LDR, usually the discriminating probe is labeled with Cy3 and the common probe is linked with a cZip tail at the 3 end. Our previous work [13] and this study showed that labeling common probe with Cy3 and linking discriminating probe with cZip are also effective for detecting PCR products. In multiplex PCR coupled with the LDR, the use of discriminating probe and common probe specific for each target sequence highly increases the specificity for detecting the amplicons [12,13,15,16]. However, using LDR to verify PCR products sometimes may lead to false negative results due to small deviation of PCR products, especially caused by the use of Taq. In our universal array system, Zip probes fixed on the chip are complementary to cZip. These Zip sequences have the same or similar Tm values and have no relationship with the sequences of detected genes. Compared with gene-specific DNA microarray, universal array is easier to be set up and the chip can be used for detecting any exogenous genes in GM crops. During the screening and detection of GMOs, certified reference materials that are compatible with detection methods are needed for quantitative analysis of GMOs. Although we have set up internal controls (rice actin and 18S rRNA) in multiplex PCR system and negative/positive controls in universal microarray to avoid false negative/positive results, we didn’t quantitatively analyze the exogenous genes in transgenic rice due to the lack of commercially available certified reference materials for transgenic rice.In the ISAAA data base, there are only 7 transgenic rice which have been approved, including two developed by Chinese scientist, Huahui No. 1 and Bt Shanyou 63. Although the Ministry of Agriculture of China has granted safety certificates to two transgenic varieties of rice in 2009, this transgenic rice has not been approved for commercialized growing. Many new lines of transgenic rice have been developed. Our multiplex PCR-LDR-UA system can reliably detect and identify the genetically modified rice, to determine if unauthorized genetically modified rice flows to the market, and set the foundation for food safety assessment in China.
Acknowledgements
We would like to thank Dr. Guoying Xiao at Institute of Subtropical Agriculture, Chinese Academy of Sciences, for providing Bar transgenic rice seeds; Dr. Qian Qian at China National Rice Research Institute for providing Bt/Cpt I transgenic rice seeds; Dr. Shouyi Chen at Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, for providing Xa21 and OsVTE1 transgenic rice seeds; Dr. Bizeng Mao at College of Agriculture & Biotechnology, Zhejiang University, for providing RCH10 transgenic rice seeds; Dr. Qiaoquan Liu at Agricultural College of Yangzhou University for providing Lrp transgenic rice seeds. This work was supported by grants from the Science and Technology Service Network Initiative Project (KFJ-EW-STS-099, KFJ-EW-STS-031) and the Knowledge Innovation Project (KSCX2-YW-N-034) of the Chinese Academy of Sciences, the Ministry of Science and Technology of China (2012BAK01B00), and the Science & Technology Commission of Shanghai Municipality (06JC14077, 13JC1404003).References
- Toriyama K, Arimoto Y, Uchimiya H, Hinata K (1988) Transgenic rice plants after direct gene transfer into protoplasts. Nature Biotechnol 6: 1072-1074.
- Zhang W, Wu R (1988) Efficient regeneration of transgenic plants from rice protoplasts and correctly regulated expression of the foreign gene in the plants. Theor Appl Genet 76: 835-840.
- Zhang HM, Yang H, Rech EL, Golds TJ, Davis AS, et al. (1988) Transgenic rice plants produced by electroporation-mediated plasmid uptake into protoplasts. Plant Cell Rep 7: 379-384.
- Jain RK, Jain S (2000) Transgenic strategies for genetic improvement of basmati rice. Indian J Exp Biol 38: 6-17.
- Helliwell EE, Yang Y (2013) Molecular strategies to improve rice disease resistance. Methods Mol Biol 956: 285-309.
- Zimmermann A, Hemmer W, Liniger M, Luthy J, Pauli U (1998) A sensitive detection method for genetically modified MaisGardTM corn using a nested PCR-system. LWT - Food Sci Technol 31: 664-667.
- Terry CF, Harris N (2001) Event-specific detection of roundup ready soya using two different real time PCR detection chemistries. Eur Food Res Technol 213: 425-431.
- Permingeat HR, Reggiardo MI, Vallejos RH (2002) Detection and quantification of transgenes in grains by multiplex and real-time PCR. J Agric Food Chem 50: 4431-4436.
- Nadal A, Esteve T, Pla M (2009) Multiplex polymerase chain reaction-capillary gel electrophoresis: a promising tool for GMO screening--assay for simultaneous detection of five genetically modified cotton events and species. J AOAC Int 92: 765-772.
- Rudi K, Rud I, Holck A (2003) A novel multiplex quantitative DNA array based PCR (MQDA-PCR) for quantification of transgenic maize in food and feed. Nucleic Acids Res 31: e62.
- Xu J, Zhu S, Miao H, Huang W, Qiu M, et al. (2007) Event-specific detection of seven genetically modified soybean and maizes using multiplex-PCR coupled with oligonucleotide microarray. J Agric Food Chem 55: 5575-5579.
- Gerry NP, Witowski NE, Day J, Hammer RP, Barany G, et al. (1999) Universal DNA microarray method for multiplex detection of low abundance point mutations. J Mol Biol 292: 251-262.
- Qin S, Zhao X, Pan Y, Liu J, Feng G, et al. (2005) An association study of the N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) and NR2B subunit gene (GRIN2B) in schizophrenia with universal DNA microarray. Eur J Hum Genet 13: 807-814.
- Busti E, Bordoni R, Castiglioni B, Monciardini P, Sosio M, et al. (2002) Bacterial discrimination by means of a universal array approach mediated by LDR (ligase detection reaction). BMC Microbiol 2: 27.
- Bordoni R, Germini A, Mezzelani A, Marchelli R, De Bellis G (2005) A microarray platform for parallel detection of five transgenic events in foods: a combined polymerase chain reaction-ligation detection reaction-universal array method. J Agric Food Chem 53: 912-918.
- Peano C, Bordoni R, Gulli M, Mezzelani A, Samson MC, et al. (2005) Multiplex polymerase chain reaction and ligation detection reaction/universal array technology for the traceability of genetically modified organisms in foods. Anal Biochem 346: 90-100.
- Zhou PP, Zhang JZ, You YH, Wu YN (2008) Detection of genetically modified crops by combination of multiplex PCR and low-density DNA microarray. Biomed Environ Sci 21: 53-62.
- Leimanis S, Hernández M, Fernández S, Boyer F, Burns M, et al. (2006) A microarray-based detection system for genetically modified (GM) food ingredients. Plant Mol Biol 61: 123-139.