Journal of Gene Therapy
cis-Antisense RNA and Transcriptional Interference: Coupled Layers of Gene Regulation
Colleen M. Courtney and Anushree Chatterjee*
- Department of Chemical and Biological Engineering, University of Colorado at Boulder
*Address for Correspondence: Anushree Chatterjee, Department of Chemical and Biological Engineering, 3415 Colorado Avenue, UCB 596 Boulder, CO 80303, USA, E-mail: chatterjee@colorado.edu
Citation: Courtney CM, Chatterjee A. cis-Antisense RNA and Transcriptional Interference: Coupled Layers of Gene Regulation. J Gene Ther 2014; 2(1): 9
Copyright © 2014 Courtney CM, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Gene Therapy | ISSN: 2381-3326 | Volume: 1, Issue: 1
Submission: 14 December 2013 | Accepted: 28 January 2014 | Published: 05 February 2014
Abstract
Antisense transcription is omnipresent occurring broadly in most living organisms. Growing evidence suggests the presence of noncoding cis-antisense RNA’s that can silence gene expression. Recent studies also indicate the role of transcriptional interference in regulating expression of neighboring genes arranged in convergent orientation. A combination of transcriptional interference and cis-antisense RNA interaction has the potential to add multiple-levels of regulation which can allow such a system to have a tunable and complex higher-order system response to environmental stimuli. This presents an important insight into the functional role of antisense transcription.
Keywords
Antisense transcription, Convergent Promoters, Antisense RNA, Transcriptional Interference, RNA Polymerase, RNA Polymerase Collision
Antisense Transcription: A Widespread Occurrence in Genomes
Proteins which regulate gene expression have been studied in great detail; however, only recently RNA is coming to light as a key regulatory molecule that controls gene expression [1]. The many pathways in which RNA can regulate gene expression include non-coding RNAs which cause epigenetic modifications [2], RNAs which interact with proteins to alter gene expression such as the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas system [3,4], RNA interference in eukaryotes [5], and direct interaction between complementary antisense RNAs [6,7] that modify expression of genes participating in various cellular processes including physiological responses, housekeeping functions, metabolism, and pathogenic processes [1,7,8]. In particular, with the recent advent of RNA-sequencing technologies and tiling arrays, a large number of sense-antisense RNA transcripts have been reported in both prokaryotic [9-15] and eukaryotic genomes [16-19]. Approximately thousands of antisense gene pairs have been found in the human genome, many thought to be involved in life-threatening diseases including breast cancer [20], pancreatic cancer [21,22] and HIV [23]. Until recently bulk of bacterial genomes were considered to consist of protein-coding regions, however, this picture is changing drastically with exponential increase in identification of cis-antisense RNA in a range of bacteria and archea, including, Escherichia coli [9], Salmonella enterica [24], Mycoplasma pneumonae [14], Synechocystis sp. PCC 6803 [25], Lysteria spp. [13], Bacillus subtilis [26], Vibrio cholerea [12], Chlamydia trachomatis [27], Psuedomonas aeruginosa [28], Psuedomonas syringae [29], Staphylococcus aureus [30], and Sinorhizobium meliloti [31]. Increasing knowledge and information of abundance of antisense genes has caused speculation that antisense transcription is an important hidden layer of regulation [16,32-34].
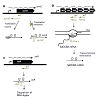
A pair of genes are said to be antisense to each other, when they are present on opposite strands of DNA (one on sense and other on the antisense strand), with corresponding promoters convergent to each other, such that there is a partial overlap between transcripts [Figure 1a-c]. Such convergent transcription results in production of complementary transcripts, also known as cisencoded sense-antisense transcripts or naturally occurring antisense RNAs (asRNAs) [34]. Two main mechanisms have been reported to operate among such sense and antisense transcripts, namely, transcriptional interference and antisense RNA interactions [1,35,36]. Transcriptional interference is defined as the suppressive influence of one transcriptional process on an adjacent transcriptional process occurring in cis due to RNA polymerase (RNAP) traffic along the DNA [35] and has been reported in a number of studies in both prokaryotic [32,37-39] and eukaryotic systems [33,40,41]. On the other hand, cis-encoded antisense RNAs generated from opposite strands of the DNA have the potential to form extensive base-pairing interactions with corresponding sense RNAs [42,43] and target them for either translational inhibition (Figure 1a), transcriptional attenuation (Figure 1b), or RNA degradation (Figure 1c) [36,44-46].
Figure 1: RNA regulatory mechanisms during antisense transcription: Sense and antisense RNA are indicated in black and grey respectively, the black block arrows represent protein coding regions/open reading frames (orf), 5′ UTRs are the regions upstream of the orf. Three regulatory mechanisms are shown. (a) Translational inhibition: In Escherichia coli, binding of asRNA symR to symE blocks the RBS of symE mRNA, preventing production of toxin-like endonuclease symE. (b) Transcriptional attenuation: In Vibrio anguillarum binding of RNAβ to a nascent fatDCBA-angRT transcript induces premature termination after fatA gene, resulting in high expression of fatDCBA mRNA and low expression of angRT mRNA. (c) RNA degradation or cleavage: In Synechocystis sp. PCC 6803, binding of asRNA isiR to isiA mRNA induces degradation of the duplex
Much of the work on antisense transcription focuses either on the role of transcriptional interference alone or antisense interaction alone. Models have been created to individually characterize the effects of antisense interactions [1,7,46] or transcriptional interference [38,47,48] but little work has been reported on combined interference studies [32]. In this review, we look at both these mechanisms, and consider the prospect of higher order system behavior when both of these mechanisms occur simultaneously.
Antisense RNAs and RNA Interaction Mechanisms in Bacteria
cis-Antisense RNA is produced when transcription occurs from the DNA strand opposite to a transcriptional unit (Figure 1a-c). cis-Antisense RNA’s tend to overlap either at the 5′ end (head to head overlap), such as the prgQ/prgX gene pair of E. faecalis [32], MgtC/AmgR sense antisense pair in S. enterica [49], hok/sok toxin-antitoxin system in E. coli [50]; or at the 3′end (tail to tail overlap), such as in the case of alr1690, which overlaps the adjacent gene all1691 gene encoding the ferric uptake regulator in Cyanobacterium Anabaena sp. PCC 7120 [51] and tpxA/ratA sense-antisense pair in B. subtilis [8]. cis-antisense RNAs can exist is various sizes in naturally occurring systems, ranging between short antisense RNAs, such as the 69 nt Sar RNA of bacteriophage 22 [52], 77 nt symR RNA of E. coli [53], 77 nt OOP RNA of bacteriophage λ [54], and 104 nt Anti-Q RNA of E. faecalis [32], and long antisense RNA’s, such as the 1200 nt AmgR asRNA of S. enterica, 2 kb Anti2095 RNA of Lysteria monocytogenes [13], and 7kb MED4 RNA of Prochlorococcus spp. [55].
Similar to proteins, in order to be functional RNA molecules require specific secondary and tertiary structures [42,46,56,57]. Frequently, interaction between two or more RNA molecules is catalyzed via single stranded regions such as hairpins, stem loops and bulges [58]. Typically, binding of sense/antisense RNA can cause three kinds of outcomes: (i) translational inhibition due to blocking of the ribosome binding site [53], (ii) RNA degradation due to action of RNases (RNAses III, E, etc.) [46,59], and (iii) transcriptional attenuation due to structural changes which destabilize RNAP:RNA complex and consequently terminate transcription [60]. Translational inhibition is exemplified by the regulation of symE mRNA, encoding the toxin-like endonuclease symE in E. coli, by the asRNA symR [53]. The symE/symR transcripts overlap at the 5′ end, and include the ribosomal binding site (RBS) and start site of symE (Figure 1a). The symE/symR duplex results in blocking of the RBS of symE, thus preventing translation of symE transcript. Similarly, in S. auerus binding of asRNA sprAIAS to the Shine-Delgarno sequence and AUG start site of sprAI mRNA prevents the translation of sprAI mRNA, thus inhibiting expression of the toxin SprAI [61]. A similar mechanism is shared by a number of Type I toxin-antitoxin systems, including hok/sok gene pair in E. coli, tpxA/ratA gene pair in B. subtilis and RNA I/RNA II systems in E. faecalis [8].
Transcriptional attenuation is exemplified by interaction of RNAβ with the fatDCBA mRNA to induce transcriptional termination after the fatA gene in the fatDCBA-angRT operon in Vibrio anguillarum [62] (Figure 1b). This results in high levels of expression of fatDCBA mRNA, and consequently low levels of expression of angRT mRNA. In E. faecalis, interaction between the 104 nt short asRNA Anti-Q produced from the prgX operon and the complementary prgQ mRNA prevents elongation of the nascent prgQ transcript past a putative terminator, causing premature termination of prgQ transcript via inhibition of anti-terminator formation [60].
Regulation of RNA stability due to antisense interaction is exemplified by the asRNA gadY which binds to polycistronic gadXW transcript to induce RNaseIII mediated cleavage and release of monocistronic gadX and gadW transcripts [63]. Similarly, the 77nt OOP RNA of λ phage interacts with CII mRNA and targets it for degradation via RNaseIII-dependent cleavage, thus preventing production of the CII repressor [54]. The isiA/isiR sense-antisense pairs in Synechocystis sp PCC 6803 form a duplex, which causes degradation of the isiR mRNA, though via an unknown mechanism [64] (Figure 1C). On the other hand, binding of MED4 asRNA to polycistronic complementary RNA in fact protects the latter from RNaseE mediated cleavage by protecting the RNAseE recognition sites, thereby affording stability to the polycistronic mRNA [55]. Similarly, in E. faecalis the interaction between QS RNA, produced from the prgQ operon, and the complementary prgX transcript, causes RNaseIII-dependent cleavage of 5′UTR of the prgX mRNA which in turn enhances translation of the prgX mRNA [65].
Transcriptional Interference: Mechanisms and Switch Response
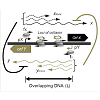
Transcriptional interference occurs when one transcriptional process suppresses an adjacent transcriptional process due to RNAP traffic along the DNA [35] and has been reported in both prokaryotic [37-39] and eukaryotic systems [33,40,41,66]. Transcriptional interference utilizes RNAP traffic to control gene expression and serves as a short-cut to gene regulation as it can interfere with transcriptional initiation, elongation as well as termination [35]. Transcriptional interference can occur via four mechanisms: (i) RNA polymerase collision, whereby elongating RNA polymerase fired from both the promoters collide with each other (Figure 2a), (ii) sitting Duck model, in which an elongating RNAP collides with a stationary RNAP (Figure 2b), (iii) road block model, where a DNA bound protein complex hinders RNAP movement along the DNA (Figure 2c), and (iv) occlusion model, where movement or binding of RNAP at one of the promoters blocks RNAP from binding at the other promoter (Figure 2d), potentially also causing competition for activators [35].
Figure 2: Mechanisms of transcriptional interference. Schematic of a general system of convergent promoters pX and pY is shown. As an example pX is considered to be the aggressive promoter. Four modes of transcriptional interference are shown. (a) RNAP collision, converging RNAPs collide within the overlapping DNA. (b) Sitting duck collision, an elongating RNAP from pX collides with a stationary RNAP at the weaker pY promoter. (c) Roadblock, DNA bound protein complex proximal to pY hinders an elongation complex from pX. (d) Promoter occlusion, binding of RNAP at the pY promoter is hindered by elongation complex from pX (or by binding of RNAP at pX when distance between pX-pY is short, not shown here)
During convergent transcription for successful transcription to occur RNAPs need to traverse the length of overlapping DNA to form a full-length transcript. Co-transcription from such a locus either results in successful transcription where RNAPs continue elongation in absence of converging RNAPs from the opposing promoter, or failed transcription when converging RNAPs collide, causing one or both RNAPs to fall-off the DNA (Figure 3). While a significant fraction of collided RNAPs fall off the DNA, a fraction of collided RNAPs backtrack and resume movement along the DNA after a temporary stall [67]. For set of two general convergent promoters pX and pY (Figure 3), the frequency of RNAP collision due to co-transcription from both the promoters depends on multiple factors: (i) relative strengths of promoter pX and pY, (ii) length of overlapping DNA between the two promoters (the probability of RNAP collisions increases with distance), and (iii) sequence context of overlapping DNA.
Under biologically relevant conditions the relative strengths of promoters pX-pY can vary between two states, one in which pX is more aggressive than pY promoter, i.e. rate of RNAP firing from pX promoter (fX) is higher than that from pY (fY) and other in which pY promoter is more aggressive than pX (i.e. fX< fY). In state 1, the RNAPs firing from the aggressive promoter pX are more likely to succeed in making a successful x transcript and RNAP collision would be more fatAl for pY promoter with little or no production of successful y transcripts (Figure 3). When pX is the aggressive promoter, most of the collisions would occur proximal to the weaker promoter pY as has been seen for the convergent promoters pR-pL of coliphage 186 [37] and PQ-PX of pCF10 plasmid in E. faecalis [32,68]. Similarly, the opposite holds true for state 2 when pY is the more aggressive promoter. RNAP collisions would exert greater suppression on x expression in state 2, compared to y expression. This is exemplified by studies on the gal7 and gal10 genes of S. cerevisiae, where arranging genes in convergent orientation suppresses transcription from this region due to increased RNAP collision [40].
Although it was first thought that only transcriptional interference from strong promoters could affect weak promoters, both mathematical modeling [32,47,69,70] and experiments [32,37,70] suggest that minor differences in strengths of convergent promoters can give rise to significant transcriptional interference. Stochastic simulations and experimental analysis of convergent transcription in the PR-PRE promoter pair of bacteriophage λ showed a 5.5 fold change in expression from the stronger PR promoter due to interference from the weaker PRE promoter [38]. This was attributed mainly to the presence of RNAP initiation complexes at the weaker promoter, which acted as sitting ducks for collision with elongating RNAP originating from the stronger promoter, such as that seen in the pR-pL promoters of coliphage 186 [37].
The probability of RNAP collision depends on the residence time of converging RNAPs in the overlapping DNA. If the length of the overlapping DNA is short then occlusion effects are more likely [71]. For longer overlapping DNA (>>RNAP footprint), occlusion effects can be neglected and RNAP collision is the more dominant mechanism of transcriptional interference. RNAP collision is also more pronounced when the overlapping distance is increased [37] or when the velocity of RNAP decreases within the overlapping region due to presence of pause sites as reported for PR-PRE promoter pair in bacteriophage λ [38]. Both these effects can increase the net residence time of RNAP in the overlapping region, thus increasing the probability of RNAP collision.
During antisense transcription, under biologically relevant conditions where the relative strengths of promoters pX-pY vary between two states, the net effect of transcriptional interference tends to amplify the gap between expression levels of full length transcripts x and y, compared to a case if the promoters were arranged in tandem. In cases where pX and pY drive expression of genes which give rise to opposing phenotypes, transcriptional interference can serve as an important gene regulatory mechanism that can give rise to switch-like behavior. This is exemplified by the role of transcriptional interference in conferring a bistable genetic-switch behavior to the prgQ/prgX operon controlling conjugative transfer of drug-resistance plasmid pCF10 between donor and recipient cells in pathogen E. faecalis [32]. It was shown that under a “conjugationally-incompetent” or “off” state, expression from a repressed PQ promoter (driving prgQ expression) decreased 90 fold due to convergent transcription from an equally strong PX promoter (driving prgX expression). On the other hand, in a “conjugationally-incompetent” or “on” state, transcription from the PX promoter had only marginal effect on the transcription from a 10-foldstronger de-repressed PQ promoter, causing expression of the 530 nt QL RNA capable of inducing conjugation-causing genes in the donor cell. Transcriptional interference has also been shown to facilitate two distinct bistable phenotypes in infectious pathogen Bordetella bronchiseptica [72]. The interference causes two populations to exist in the lungs: Bvg+ responsible for the in vivo infectious state and Bvg responsible for survival ex vivo. The interference which causes these bistable phenotypes allows the infection to thrive in the lungs and survive to infect others.
Wreckage of RNAP collision: A Source of Antisense RNA?
During antisense transcription, collision between converging RNAPs results in premature termination of transcriptional progress of one or both elongation complexes, thus giving rise to a mixture of truncated and full length sense and antisense RNA sequences [32]. Depending on the relative firing rates of RNAP and overlapping sequence, converging RNAPs may collide at various loci along the DNA, thus giving rise to a distribution of different sizes of truncated RNA both in the sense and antisense direction (denoted by xtrunc and ytrunc in Figure 3). A less explored aspect of transcriptional interference relates to the potential regulatory role of such truncated RNA. It is plausible that truncated transcripts with a certain minimum size possess secondary structures that can cause interaction with antisense counter transcripts. Though the sequence of overlapping DNA would vary between different systems, the advantage of antisense transcription is that it allows for extensive base pairing between truncated RNA and the full-length antisense counter transcripts, hence enhancing the probability of RNA interaction. Both short antisense-RNAs [50,52-54,73,74] and long antisense RNA [49,75-77] have been shown to participate in antisense interaction in various bacterial systems. Therefore it is possible that the resulting sense, antisense RNA hybrid complexes between truncated and full-length RNA may be subjected to similar mechanisms of RNA degradation, transcriptional attenuation or translational inhibition [6,46]. The presence and functional role of truncated RNA produced as a result of RNAP collision has been shown for the prgQ/prgX operon of pCF10 plasmid in E. faecalis [32]. Under de-repressed conditions when effect of transcriptional interference is more pronounced in prgQ/prgX locus, truncated PQ and PX transcripts of sizes ranging between approximately 100-200 nt and 80-200 nt respectively are observed, all of which lie within the overlapping region of 223 bp of prgQ/prgX genes. Under de-repressed conditions when transcriptional interference effects were less pronounced, truncated RNA are less abundant. Northern analysis showed that overexpression of a 223 nt truncated PQ RNA in trans repressed expression of prgX mRNA, whereas overexpression of a 104 nt truncated PX RNA in trans repressed expression of prgQ mRNA, thus indicating that truncated RNA are capable of suppressing the expression of counter transcripts. Similarly in the ubiG/mccBA operon of Clostridium acetobutylicum truncated RNA of various sizes ranging between 200-700 nt lacking Rho-dependent terminator structures at 3′ end were found [78]. The expression of the truncated RNA was independent of RNase III and RNAse J1/J2 cleavage, which could potentially hint at RNAP collision based termination mechanism. Northern analysis of sense-antisense transcripts in higher eukaryotes such as mouse, A. thaliana indicate presence of shorter transcripts that lack poly-A tail and are nuclear localized [33]. These truncated transcripts have been found to be richer at 5′ends compared to 3′ends (poly A rich), thus indicating that these could be an outcome of transcriptional interference or local sense, antisense effects.
Figure 3: Regulatory mechanisms during antisense transcription. Schematic of a general system of convergent promoters pX and pY driving the expression of genes X and Y respectively, the black and grey block arrows represent protein coding regions/open reading frames (orf). The overlapping DNA between the pX-pY promoters is indicated by length L. RNAPs fire from pX and pY with frequency fX and fY respectively. Successful transcription results in expression of full length transcripts x and y (bold arrows) from pX and pY respectively. During RNAP collision one or both of the elongating RNAP’s fall off the DNA giving rise to different sizes of truncated RNA xtrunc and ytrunc (dashed arrows) from pX and pY respectively depending on the loci of RNAP collision. Full-length and truncated RNA share extensive base-pairing and potentially exert antisense interactions on each other.
With exception of few studies [32,69,78,79], the presence of truncated RNA has not yet been vigorously investigated in systems with antisense transcription. The plethora of cis-antisense noncoding RNAs found in bacteria could potentially be the wreckage of RNAP collision due to transcription from conditionally activated hidden promoters, thus hinting that this could be rather a ubiquitous phenomenon.
Coupled effect of Transcriptional interference and Antisense RNA interaction
The combined effect of transcriptional interference and antisense interaction between truncated and full-length sense and antisense RNA can further sharpen the switch response compared to when only one of these mechanisms exist. Transcriptional interference can potentially give rise to two-fold regulation, (i) reduction of full-length transcript levels due to RNAP collision, (ii) generation of truncated RNA capable of exerting antisense interactions on counter transcripts. Four potential combinations of transcriptional interference and antisense regulation effects are shown in (Figure 4a-d). The sharpest switching response is likely to when both these mechanisms occur simultaneously (Figure 4e-f). Depending on the relative strength of promoters pX-pY, the loci of collision will shift towards the weaker promoter. If we consider pX is the stronger promoter, collisions would occur near the pY promoter, thus the truncated RNA from pY would be very short and unlikely to interact with sense pX RNA. In this case, majority of the truncated sense pX RNA will have nearly the length of overlapping region, and hence possess a higher potential to interact with a nascent pY transcripts. Therefore, even if a nascent pY transcript escapes RNAP collision, it would still be swamped by the relatively large pool of truncated sense pX RNA (Figure 4e). The relative stoichiometry of sense and antisense would influence the final extent of suppression. The situation would be reversed when pY becomes the stronger promoter (Figure 4f).
Antisense transcription can result in complex cellular behavior, especially in context of a biological gene network. Since antisense transcription can amplify the gap between transcript expression between two physiologically different states, such a gene regulatory mechanism is capable of showing reciprocal switch like behaviors such as bistable switch response in prgQ/prgX operon of E. faecalis [32,79,80] and scbA/scbR operon of S. coelicolor [69]. Antisense transcription from the prgQ/prgX locus of conjugative plasmid pCF10 of E. faecalis, allows for controlling the expression of long prgQ mRNA, which induces expression of downstream conjugation causing genes.
Transcriptional interference from downstream PX promoter, as well as, antisense RNA interaction exerted by a 104 nt non-coding Anti-Q RNA expressed from the PX promoter, causes premature termination of a nascent prgQ transcript, thus preventing conjugative transfer of pCF10 plasmid. Both experiments and mathematical modeling showed that for this system bistable switch behavior was only observed when both mechanisms of transcriptional interference and antisense interaction operate simultaneously. Using mathematical modeling it was shown that antisense transcription confers a bistable switch to the scbA/scbR gene pair of S. coelicolor, which allows regulation of expression of scbA mRNA, which encodes the key enzyme ScbA involved in synthesis of γ-butyrolactones that regulate antibiotic biosynthesis in the S. coelicolor [69].
In ubiG/mccBA operon of C. acetobutlyicum, both mechanisms of transcriptional interference and RNA interaction confer a genetic switch regulating the expression of ubiG operon, which contains genes required for conversion of methionine to cysteine [78]. In presence of methionine, transcription from the stronger T-box promoter causes premature termination of the antisense S-box transcripts. As a result, the levels of S-box riboswitch antisense RNA decreases, which in turn increases the expression of full length ubiG mRNA, which encodes enzymes required for conversion of methionine to cysteine. On the other hand, under conditions of high levels of cysteine, transcription from the downstream S-box promoter tends to reduce the expression of ubiG mRNA. Similarly, antisense transcription from the icsA/RnaG locus of virulence plasmid pINV of Shigella flexneri, allows controlling the expression of icsA mRNA, which encodes an invasion protein required for colonization of host by the bacterial pathogen [81]. This locus encodes a non-coding antisense RnaG RNA, which overlaps with icsA mRNA at the 5′ end, and has been shown to cause premature termination of icsA mRNA following a transcriptional attenuation mechanism [81]. In addition to antisense RNA interaction mediated regulation, the stronger RnaG promoter exerts transcriptional interference on the weaker icsA promoter icsA, further reducing activity of the latter.
Outlook: Antisense Transcription a Widespread Mechanism of Gene Regulation
Antisense transcription is omnipresent in bacteria, archaea, and eukaryotic genomes. One could argue that shorter prokaryotic genomes use antisense transcription for conserving space, however presence of thousands of such cis-antisense gene pairs in relatively larger eukaryotic genomes [16-19] clearly refutes such an argument and points towards potential role of antisense transcription as a mechanism of gene regulation conserved over evolution [82]. A large fraction of mechanistic studies on antisense transcription have been performed in prokaryotic systems which are characterized by shorter intergenic distances [32,37-81,46]. There are many systems yet to be characterized that hypothetically will exhibit both antisense interactions and transcriptional interference. The large number of cis-oriented promoters found in bacteria, yeast, flies, HIV, and mouse [48] may lead one to speculate why these systems are so prevalent in nature and what is their role in gene regulation and phenotype determination.
Antisense transcription may have a more pronounced effect in systems with a longer overlapping region, found commonly in mammalian genomes [16]. It is interesting to note that in a large fraction of convergent promoter based gene pairs in mammalian genomes, often one out of the two genes tends to express non-coding RNA [83], potentially opposing the coding RNA. In many cases such convergent transcription gives rise to reciprocally regulated switch [33,83]. Bioinformatics techniques are being used to identify cis-antisense pairs in order to characterize more of these systems in diverse species. Using bioinformatics, the prevalence of cis-oriented genes has been reported to be: 26.3% in humans [17,84,85], 21.9% in mice [16,84,86,87], 16.8% in drosophila [18,88], 2.8% in C. elegans, 15.8% in sea squirt 6.6% in chickens, 4.5% in rats, 4.3% in frogs, 2.2% in zebrafish, 3.8% in cows [84], and 8.9% in Arabidopsis [88-90]. While bioinformatics has been useful to identify cis-antisense loci, the extent of the activity of these non-coding RNAs and the exact function of most sense, antisense pairs remains to be determined [34,91]. Moreover, a concerted effort is required to examine these systems for antisense interactions and transcriptional interference to determine their combined role in regulating gene expression levels and phenotype determination.
Figure 4: Coupled effect of antisense RNA interaction (AI) and transcriptional interference (TI) during antisense transcription. (a-d) Schematic showing four possible combinations of mechanisms of transcriptional interference (TI) and Antisense interaction (AI) regulating expression from pX and pY: None (a), TI only (b), AI only (c), both TI and AI (d). (e-f) Steady state levels of full-length RNA x (e) and y (f), expressed from promoters pX and pY respectively, for various ratios of fY/fX for the four cases considered in a-d. For a system transitioning from one value of fY/fX to other, maximum switching response occurs when both TI and AI effects are present.
In this review, we highlight the regulatory advantage that cells can achieve via coupled role of transcriptional interference and cis-asRNA based regulation during antisense transcription. Importantly, antisense transcription offers a number of regulatory advantages over trans-encoded asRNAs and regulatory proteins. Firstly, cis-antisense RNAs share extensive base pairing with their cognate target RNAs leading to more effective antisense interactions, compared to transasRNAs which have only partial complementarity with their target RNAs. Further, during antisense transcription due to coupled activity of both convergent promoters as well as localized RNA expression, cis-asRNA are more likely to have a faster kinetic effect compared to trans asRNA or regulatory proteins. In general, regulatory proteins take a longer time to act since both steps of transcription and translation are required for the proteins to be functional, compared to asRNA which only require transcription. For trans-asRNA based regulation the relative stoichiometry of sense and antisense RNA influences the final extent of suppression. In contrast, during antisense transcription, regulation via cis-asRNA interactions as well repression by transcriptional interference can contribute to the gene regulation. Moreover, using a single repressor which acts on one of the promoters, antisense transcription can allow for simultaneous regulation of target RNA and cis-asRNA expression [32,38,69]. Such a coupled effect of transcriptional interference and cis-asRNA interaction tends to amplify the gap between sense and antisense RNA expression. This allows antisense transcription to act both as biological switch and potentially as a noise filter against fluctuating environmental signals. Given the plethora of sense, antisense pairs in both prokaryotic and eukaryotic genomes, the next obvious question to ask is whether sense, antisense pairs are being regulated by both transcriptional interference and antisense regulation at a genome scale. From a synthetic biology point of view, given that promoter strengths, length and sequence of overlapping DNA are highly tunable, antisense transcription could be exploited to tweak naturally existing networks or create novel networks for obtaining desired characteristics. Finally, a closer look at antisense gene pairs is needed to understand whether convergent transcription is merely an act of serendipity in nature or a well-designed set-up of gene regulation.
Acknowledgements
This work was supported by the University of Colorado start-up funds to AC. CMC was partially supported by NIH Pharmaceutical Biotechnology training grant 5T32GM008732.
References
- Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P (2013) The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol 11:75-82..
- Magistri M, Faghihi MA, St Laurent G, Wahlestedt C (2012) Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet 28: 389-396.
- Sorek R, Kunin V, Hugenholtz P (2008) CRISPR-a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6:181-186.
- Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, et al. (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67-71.
- Wilson RC, Doudna JA (2013) Molecular mechanisms of RNA interference. Annu Rev Biophys 42: 217-39.
- Brantl S (2002) Antisense-RNA regulation and RNA interference. Biochim Biophys Acta 1575:15-25.
- Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136: 615-628.
- Gerdes K, Wagner EGH (2007) RNA antitoxins. Curr Opin Microbiol 10:117-24.
- Dornenburg JE, Devita AM, Palumbo MJ, Wade JT (2010) Widespread antisense transcription in Escherichia coli. MBio 1: e00024-10. 1 5
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, et al. (2010) The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250-255.
- Han Y, Lin YB, An W, Xu J, Yang HC, et al. (2008) Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional read through. Cell Host Microbe 4:134-46.
- Liu JM, Livny J, Lawrence MS, Kimball MD, Waldor MK, et al. (2009) Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res 37: e46.
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, et al. (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950-956.
- Güell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, et al. (2009) Transcriptome Complexity in a Genome-Reduced Bacterium. Science 326:1268-1271.
- Georg J, Voss B, Scholz I, Mitschke J, Wilde A, et al. (2009) Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol 5:305.
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564-1566.
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, et al. (2003) Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol 21: 379-86.
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, et al. (2002) Annotation of the Drosophila melanogaster euchromatic genome: asystematic review. Genome Biol 3: 1-22.
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, et al. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842-846.
- Berteaux N, Aptel N, Cathala G, Genton C, Coll J, et al. (2008) A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol 28: 6731-45.
- Monti L, Cinquetti R, Guffanti A, Nicassio F, Cremona M, et al. (2009) In silico prediction and experimental validation of natural antisense transcripts in two cancer-associated regions of human chromosome 6. Int J Oncol 34: 1099-108.
- Marshall L, White RJ (2008) Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer 8: 911-914.
- Coiras M, López-Huertas MR, Pérez-Olmeda M, Alcamí J (2009) Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol 7:798-812.
- Kröger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, et al. (2012) The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109: E1277-86.
- Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, et al. (2011) An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A 108: 2124-2129.
- Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, et al. (2012) Condition-Dependent Transcriptome Reveals High-Level Regulatory. Science 335: 1103-1106.
- Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T (2010) Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome.Nucleic Acids Res 38: 868-877.
- Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, et al. (2012) The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8: e1002945.
- Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, et al. (2010) Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol 192: 2359-72.
- Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, et al. (2010) Cartography of methicillin-resistant S. aureus transcripts: detection,orientation and temporal expression during growth phase and stress conditions. PLoS One 5: e10725.
- Schlüter JP, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, et al. (2010) A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genomics 11: 245.
- Chatterjee A, Johnson CM, Shu CC, Kaznessis YN, Ramkrishna D, et al. (2011) Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc Natl Acad Sci U S A 108:9721-9726.
- Hongay CF, Grisafi PL, Galitski T, Fink GR (2006) Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127:735-45.
- Georg J, Hess WR (2011) cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75: 286-300.
- Shearwin KE, Callen BP, Egan JB (2005) Transcriptional interference--a crash course. Trends Genet 21: 339-345.
- Thomason MK, Storz G (2010) Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet 44:167-188.
- Callen BP, Shearwin KE, Egan JB (2004) Transcriptional Interference between Convergent Promoters Caused by Elongation over the Promoter. Mol Cell 14: 647-656.
- Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE (2009) Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol Cell 34: 545-55.
- Ward DF, Murray NE (1979) Convergent Transcription in Bacteriophage lambda: Interference with Gene Expression. J Mol 133: 249-266.
- Greger IH, Aranda A, Proudfoot N (2000) Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. PNAS 97: 8415-8420.
- Gullerova M, Proudfoot NJ (2008) Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132: 983-95.
- Franch T, Petersen M, Wagner EG, Jacobsen JP, Gerdes K (1999) Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J Mol Biol 294: 1115-1125.
- Bennett CF, Swayze EE (2010) RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol 50: 259-293.
- Johnson E, Srivastava R (2012) Volatility in mRNA secondary structure as a design principle for antisense. Nucleic Acids Res 41: 1-10.
- Yamaguchi Y, Park J-H, Inouye M (2011) Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet 45: 61-79.
- Brantl S (2007) Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol 10: 102-9.
- Sneppen K, Dodd IB, Shearwin KE, Palmer AC, Schubert RA, et al. (2005) A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J Mol Biol 346: 399-409.
- Palmer AC, Egan JB, Shearwin KE (2011) Transcriptional interference by RNA polymerase pausing and dislodgement of transcription factors.Transcription 2: 9-14.
- Lee EJ, Groisman EA (2010) An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol 76: 1020-1033.
- Thisted T, Gerdes K (1997) Mechanism of Post-segregational Killing of Plasmid Rl by the hok/sok System Sok Antisense RNA Regulates hok Gene Expression Indirectly Through the Overlapping mok Gene. J Mol Biol: 41-54.
- Hernández JA, Muro-Pastor AM, Flores E, Bes MT, Peleato ML, et al. (2006) Identification of a furA cis-antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J Mol Biol 355: 325-34.
- Liao SM, Wu TH, Chiang CH, Susskind MM, McClure WR (1987) Control of gene expression in bacteriophage P22 by a small antisense RNA. I.Characterization in vitro of the Psar promoter and the sar RNA transcript. Genes Dev 1: 197-203.
- Kawano M, Aravind L, Storz G (2007) An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol Microbiol 64: 738-54.
- Krinke L, Wulff DL (1987) OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependentmechanism. Genes Dev 1: 1005-1013.
- Stazic D, Lindell D, Steglich C (2011) Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res 39: 4890-4899.
- Montange RK, Batey RT (2008) Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys 37: 117-133.
- Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY (2011) Understanding the transcriptome through RNA structure. Nat Rev Genet 12: 641-655.
- Brantl S (2002) Antisense-RNA regulation and RNA interference. Biochim Biophys Acta - Gene Struct Expr 1575: 15-25.
- Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, et al. (2010) The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 34: 883-923.
- Johnson CM, Manias DA, Haemig HA, Shokeen S, Weaver KE, et al. (2010) Direct evidence for control of the pheromone-inducible prgQ operon of Enterococcus faecalis plasmid pCF10 by a counter transcript-driven attenuation mechanism. J Bacteriol 192: 1634-42.
- Sayed N, Jousselin A, Felden B (2012) A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat Struct Mol Biol 19: 105-112.
- Stork M, Di Lorenzo M, Welch TJ, Crosa JH (2007) Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J Bacteriol 189: 3479-3488.
- Opdyke JA, Fozo EM, Hemm MR, Storz G (2011) RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol 406: 29-43.
- Dühring U, Axmann IM, Hess WR, Wilde A (2006) An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci U S A 103: 7054-7058.
- Johnson CM, Haemig HH, Chatterjee A, Wei-Shou H, Weaver KE, et al. (2011) RNA-Mediated Reciprocal Regulation between Two Bacterial Operons is RNase III Dependent. MBio 2: e00189-11.
- Prescott EM, Proudfoot NJ (2002) Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci 99: 8796-8801.
- Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH (2006) Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res 34: 5416-5425.
- Shu CC, Chatterjee A, Dunny G, Hu WS, Ramkrishna D (2011) Bistability versus bimodal distributions in gene regulatory processes from population balance. PLoS Comput Biol 7: e1002140.
- Chatterjee A, Drews L, Mehra S, Takano E, Kaznessis YN, et al. (2011) Convergent transcription in the butyrolactone regulon in Streptomycescoelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS One 6: e21974.
- Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE (2009) Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol Cell 34: 545-555.
- Bendtsen KM, Erdossy J, Csiszovszki Z, Svenningsen SL, Sneppen K, et al. (2011) Direct and indirect effects in the regulation of overlapping promoters. Nucleic Acids Res 39: 6879-6885.
- Mason E, Henderson MW, Scheller EV, Byrd MS, Cotter PA (2013) Evidence for phenotypic bistability resulting from transcriptional interference of bvgAS in Bordetella bronchiseptica. Mol Microbiol 90: 716-733.
- Stougaard P, Molin S, Nordstrom K (1981) RNAs involved in copy-number control and incompatibility of. Proc Natl Acad Sci 78: 6008-6012.
- Tomizawa J, Itoh T (1981) Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci 78: 6096-6100.
- Hernández JA, Muro-Pastor AM, Flores E, Bes MT, Peleato ML, et al. (2006) Identification of a furA cis-antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J Mol Biol 355: 325-334.
- Stazic D, Lindell D, Steglich C (2011) Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res 39: 4890-4899.
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, et al. (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459: 950-956.
- André G, Even S, Putzer H, Burguière P, Croux C, et al. (2008) S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res 36: 5955-5969.
- Chatterjee A, Cook LC, Shu CC, Chen Y, Manias DA, et al. (2013) Antagonistic self-sensing and mate-sensing signaling controls antibiotic resistance transfer. Proc Natl Acad Sci U S A 110: 7086-7090.
- Cook L, Chatterjee A, Barnes A, Yarwood J, Hu WS, et al. (2011) Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol Microbiol 81: 1499-1510.
- Giangrossi M, Prosseda G, Tran CN, Brandi A, Colonna B, et al. (2010) A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri. Nucleic Acids Res 38: 3362-3375.6.
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD (2005) Genomewide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet 21: 322-336.
- Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K (2005) Disclosing hidden transcripts: mouse natural sense-antisense transcripts end to be poly(A) negative and nuclear localized. Genome Res 15: 463-74
- Zhang Y, Liu XS, Liu Q-R, Wei L (2006) Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res 34: 3465-3475.
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Chen J, et al. (2004) Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res 32: 4812–4820.
- Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y (2003) Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res 13:1324-1334.
- . Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 fulllength cDNAs. Nature 420: 563-573.
- Jin H, Vacic V, Girke T, Lonardi S, Zhu J-K (2008) Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol 9: 6.
- Jen CH, Michalopoulos I, Westhead DR, Meyer P (2005) Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol 6: R51.
- Wang XJ, Gaasterland T, Chua N-H (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6: R30.
- Sorek R, Cossart P (2010) Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 11: 9-16.