Journal of Forensic Investigation
Download PDF
Research Article
*Address for Correspondence: Simone Tredoux, Division of Forensic Medicine, Faculty of Health Sciences, University of Cape Town, 7 White Road, Plumstead, Cape Town, 7800, South Africa, Tel: +2784 325 1502; E-mail: simone_tredoux@hotmail.com
Citation: Tredoux S, Mfolozi S, Shires K. Efficiency of Buccal DNA Sampling Device in the Mortuary. J Forensic Investigation. 2015;3(2): 6.
Copyright © 2015 Tredoux S, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Forensic Investigation | ISSN: 2330-0396 | Volume: 3, Issue: 2
Submission: 31 July, 2015 | Accepted: 26 August, 2015 | Published: 31 August, 2015
Efficiency of Buccal DNA Sampling Device in the Mortuary
Simone Tredoux1*, Sipho Mfolozi1 and Karen Shires2
- 1Division of Forensic Medicine, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
- 2Division of Haematology, Department of Pathology, University of Cape Town and National Health Laboratory Services- Groote Schuur Hospital, Cape Town, South Africa
*Address for Correspondence: Simone Tredoux, Division of Forensic Medicine, Faculty of Health Sciences, University of Cape Town, 7 White Road, Plumstead, Cape Town, 7800, South Africa, Tel: +2784 325 1502; E-mail: simone_tredoux@hotmail.com
Citation: Tredoux S, Mfolozi S, Shires K. Efficiency of Buccal DNA Sampling Device in the Mortuary. J Forensic Investigation. 2015;3(2): 6.
Copyright © 2015 Tredoux S, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Forensic Investigation | ISSN: 2330-0396 | Volume: 3, Issue: 2
Submission: 31 July, 2015 | Accepted: 26 August, 2015 | Published: 31 August, 2015
Abstract
Identification of forensic DNA samples by short tandem repeat (STR) profiling is currently an essential component of criminal investigations and can aid in linking perpetrators to crimes as well as identifying missing individuals or unidentified remains. In South Africa, recent amendments to legislation have allowed for the mandatory acquisition of reference DNA samples from certain offenders in order to populate the new National Forensic DNA Database. A novel method for the collection of buccal samples, the EasiCollect device, has been proposed to facilitate the collection of these DNA samples, replacing blood collecting devices as the standard method of DNA collection. Subsequently, this device has been introduced into South African state mortuaries to assist in the identification of deceased individuals. In order to ascertain if this device is suitable for use in the postmortem setting, an investigation was performed to compare the main methodology currently utilised within South African mortuaries, namely femoral blood transferred to ‘Fast Technology for Analysis of nucleic acids’ (FTA) cards, and buccal cells obtained using the EasiCollect device. DNA yields and STR genotyping results were compared between the two collection methods in thirty deceased individuals. Buccal samples provided a significantly greater DNA yield than blood samples, while no significant difference was observed between the qualities of the sample types as measured by the 260/280 nm ratio. Full STR profiles were obtained from all blood and buccal samples, with amplification efficiency showing limited DNA degradation and PCR inhibition in these samples, and only 3% of samples giving potentially disputable results. Numerous issues surrounding the collection of blood samples, however, indicated that this method is not optimal for use in the mortuary, with the EasiCollect device providing a more practical and robust method for the collection of DNA samples in the mortuary.Keywords
EasiCollect; STR typing; Buccal cells; Blood; Decedents; DNA extractionAbbreviations
DNA: Deoxyribonucleic acid; FTA: Fast Technology for Analysis of nucleic acids; PCR: Polymerase Chain Reaction; RFU: Relative Fluorescence Unit; SAPS: South African Police Service; STR: Short Tandem RepeatIntroduction
Deoxyribonucleic acid (DNA) profiling using short tandem repeats (STRs) is widely used in the identification of forensic samples [1,2], and can aid in developing investigative leads through comparative database searches of crime scene to reference profiles [3,4]. The South African forensic DNA database currently contains a relatively small number of DNA profiles (~132 000) [5], which ultimately reduces its effectiveness as a crime solving tool. In order to populate the national database, legislation has recently been amended, now mandating the acquisition of reference DNA samples from arrested individuals and convicted offenders [6]. Following these legislative changes, there is a great need for a simple and efficient method for the collection of reference DNA samples.Blood obtained by venipuncture or finger-prick had previously been the standard method for obtaining DNA samples from living individuals [7,8]. The collection of this specimen is however invasive and requires the assistance of trained medical personnel [7,9,10]. More recently, increasing focus has been placed on buccal epithelial cells as an alternative DNA source [8-15], offering an easy-to-collect, non-invasive method [7,10-12,16,17] with no reliance on medical staff [9,10,12]. Various methods have been described for the collection of buccal cells, including saliva [11,16,17], mouthwash [8,13,14,17], cytobrushes [13,14,17] and swabs [8-11,15]. Numerous studies have reported on the differences in DNA yield and quality between these collection methods. While mouthwash samples have consistently been found to produce the highest yields of DNA when compared to other sample types [8,13,14,17],significant quantities of bacterial DNA have been observed in these samples [13,14]. Direct methods of buccal cell collection, such as cytobrushes and cotton or nylon swabs, are favourable due to the lack of a transfer step during the collection process. Buccal cells are collected directly onto these devices, thereby increasing the yields of DNA obtained using these methodologies [10]. Large amounts of bacterial contamination are however detected [9,14,15] due to the lack of DNA stabilisation within these devices. The use of foam swabs and subsequent transfer to ‘Fast Technology for Analysis of nucleic acids’ (FTA) cards provides an effective method for the collection and storage of buccal samples in which the sample bound to the card is protected against bacterial and fungal growth and enzymatic degradation [9,18]. The genotyping success rates are consistently high with this approach [11,15,19,20] and consequently the EasiCollect device (Manta Forensic, South Africa), a combination swab-FTA device, has been proposed for use as the new standardised method for the collection of reference samples from living individuals in South Africa [21].
Identification of deceased individuals by STR genotyping is inherently not an easy task, due to the lack of suitable samples for DNA extraction. Although blood was previously regarded as the standard specimen for the collection of DNA samples within the mortuary, Hansen et al. found that it is not always the best specimen for use in this setting due to the potential presence of coagulation and haemolysis which may inhibit sample collection [22]. Additional issues arise with the collection of this sample type in South African mortuaries due to the lack of a pre-packaged, sterile and quality controlled kit for sample collection. The EasiCollect device has thus also recently been introduced into state mortuaries to assist with the collection of DNA from deceased individuals.
While the DNA from buccal epithelial cells obtained by the EasiCollect device in living individuals has been shown to be comparable to extracted DNA from blood in terms of genotyping success [18], issues regarding potential DNA loss have been reported due to the non-uniform transfer from the swab to the FTA card and the small area of the card that is ultimately used for DNA extraction [10,12,15]. Additional issues may occur when using this device to collect samples from deceased individuals. These include the presence of unknown substances within the oral cavity which may act as polymerase chain reaction (PCR) inhibitors; microbial activity which may lead to rapid DNA fragmentation prior to collection; and rigor mortis of the jaw which may restrict sample collection. While the use of this device has been thoroughly tested in living subjects, its use in the mortuary setting has not been studied and it is uncertain if this is the most suitable method in this environment.
This study investigates the efficiency of the EasiCollect device in the collection of suitable DNA samples from deceased individuals. The approach was to compare the two protocols currently used for the collection of reference DNA samples within mortuaries in South Africa, namely femoral blood transferred to FTA cards and buccal epithelial cells obtained using the EasiCollect device. The quantity, quality and genotyping success of DNA obtained from each method was evaluated, as well as the practical use of each method during postmortem examinations.
Materials and Methods
Study populationSamples were collected from thirty deceased individuals during routine post-mortem examination at the Salt River Forensic Pathology Laboratory in Cape Town, South Africa. Exclusion criteria were the presence of blood, gastric contents or unknown substances in the oral cavity, bodies that were charred or decomposed, as well as infants younger than 6 months. All components of the study were approved by the Faculty of Health Sciences Human Research Ethics Committee (HREC REF: 190/2014) and consent was obtained for the collection of human tissue samples at the mortuary.
Sample collection
Blood samples: Approximately 3 ml of blood was collected from the femoral vessels of the deceased individual using a 5 ml syringe, by a trained forensic pathology officer. Around 1 ml of blood was then spotted onto each of the two designated areas on the FTA® Mini Card (non-indicating) from Whatman (UK) and allowed to dry at room temperature before returning the card to the 6x11 cm brown paper envelope.
Buccal samples: Buccal cell samples were collected from the same individuals using the EasiCollect device according to the manufacturer’s instructions (Manta Forensic, South Africa). Briefly, the sterile foam-tipped applicator of the EasiCollect device was firmly the indicating FTA card, allowing for the transfer of buccal cells from the applicator to the attached card. The handle was then bent back to allow the sample to dry, and the closed device was immediately placed into the sterile foil exhibit bag along with a desiccant.
DNA extraction and quantification
Extraction of DNA from the FTA cards was performed by Unistel Medical Laboratories (Stellenbosch, South Africa) using the Maxwell® 16 Cell LEV DNA Purification Kit (Promega, USA) and the Maxwell® 16 Instrument (Promega, USA). Buccal and blood DNA samples were eluted into 25 or 35 μl of elution buffer respectively (according to validated methodology of the Unistel Medical Laboratories).
A NanoDrop® ND1000 Spectrophotometer (Thermo Scientific, USA) was initially used to determine the concentration and purity of all extracted DNA samples. Where concentration readings fell below 10 ng/μl, DNA was re-quantified using the QubitTM dsDNA HS Assay Kit (Life Technologies, USA), and a Qubit® 2.0 Fluorometer (Life Technologies, USA) according to the manufacturer’s instructions. The total DNA yields for blood and buccal samples were calculated by multiplying the resultant concentration by the final volume of DNA extract. The purity of DNA, as assessed by spectrophotometric analysis, was measured using the 260/280 nm ratio, which measures contamination of the nucleic acid sample by protein, where a ratio of 1.8 is considered as purified DNA [23]. Quantitative PCR (qPCR) was performed on a representative subset of buccal DNA samples using the Investigator® Quantiplex Kit (Qiagen, Germany) and the Rotor- Gene 6000 (Qiagen, Germany) according to the manufacturer’s instructions.
STR genotyping
DNA profiling of the extracted DNA samples was performed by Unistel Medical Laboratories (Stellenbosch, South Africa) using a validated modification of the PowerPlex® 16 System protocol (Promega, USA). The PCR Master Mix consisted of 4 μl nucleasefree water (Promega, USA), 1 μl PowerPlex® 16 10X Primer Pair Mix (Promega, USA) and 5 μl 2X KAPA Blood PCR Mix A (KAPA Biosystems, USA) per reaction; after which 2 μl (>1 ng DNA) of isolated DNA was added to each reaction tube for a total reaction volume of 12 μl. The GeneAmp® PCR System 9700 thermal cycler (Applied Biosystems, USA) was used for the amplification of isolated DNA according to the recommendations of the PowerPlex® 16 protocol. Amplified DNA fragments were diluted into a denaturing HiDiTM formamide-internal lane standard mix and separated with capillary electrophoresis using the 3500 Genetic Analyzer (Applied Biosystems, USA). Data Analysis was subsequently performed using GeneMapper® ID-X Software.
Statistical analysis
All statistical analyses were performed using STATA version 12.0, statistical software (StataCorp LP, USA). The differences in concentration, purity and peak heights between methods of collection were compared using the two sample t-test for parametric data or the Wilcoxon’s rank-sum test for non-parametric data. Cohen’s Kappa statistic was used to determine the measurement of agreement between the alleles produced from blood and buccal samples for each individual.
Results
The quantity and quality, as well as STR genotyping performance was assessed using DNA extracted from buccal and blood samples from thirty deceased individuals from the local state mortuary facility. This was performed to ascertain the effectiveness of the EasiCollect device in collecting suitable DNA samples for human identification in this environment.DNA yield and purity
In practice, STR DNA profiling can be performed on DNA extracted from a portion of the FTA spot (3 mm punch or whole spot) or directly using an untreated 3 mm punch in the PCR reaction [24]. For this study, to be able to analyse the DNA yield, the total yield of DNA obtained from each sample was calculated as the quantity of DNA extracted from a full FTA spot, which was then used to determine the theoretical yield from a single 3 mm punch.
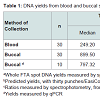
Using a combination of spectrophotometry and fluorometry to quantify the total DNA extracted in each sample, buccal DNA extracts produced DNA yields that were significantly higher than those seen in blood samples, as can be seen in Figure 1 and Table 1. The median total DNA yields obtained from a single punch was 5.4 fold higher using the EasiCollect device compared to blood drawn from the femoral vein and spotted onto the FTA card (Table 1). Figure 1 shows the distribution of calculated DNA yields per punch, which showed a highly significant (p<0.001) greater yield in buccal samples, but with both sample types showing huge yield variations: 1.57-77.53 ng in blood samples and 2.97-104.88 ng in buccal samples. Of importance is that both methods produced sufficient DNA for use in STR profiling (>1 ng/3 mm punch).
As the buccal samples in this study were thought to contain significant bacterial contamination, qPCR was performed on a subset of these samples in order to specifically quantify the yields of human DNA. As seen in Table 1, no significant difference was found between the median yield of human DNA and that of total DNA from a wholespot (p=0.2899), indicating that limited bacterial DNA was present in these samples. As a result of these findings, it was concluded that DNA yields from buccal samples were in fact significantly greater than those of corresponding blood samples collected in this study.
The purity of DNA within the samples, as measured by the 260/280 nm ratios, was used as a crude measure of DNA quality, the values for which can be seen in Table 1. No significant difference was found when comparing the 260/280 nm ratio between blood and buccal samples (p=0.2439). Notably, both sample types produced relatively poor quality DNA, with all 260/280 nm ratios being <1.7 and mean values of average ratios of 1.3 and 1.1 for blood and buccal samples respectively. This finding is a reflection of the inability of the Maxwell 16 Cell LEV DNA Purification Kit (Promega) to effectively purify DNA from the FTA substrate.
STR profile analysis
All samples in this study produced amplicons for all STR loci investigated, generating corresponding DNA profiles betweenthe blood and buccal DNA samples of each individual. The results of Cohen’s kappa test showed 100% agreement between the corresponding alleles at all 16 loci, Kappa = 1 (p<0.001).
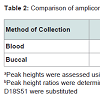
The success and reliability of the profile depends upon the uniform amplification of the various alleles and the amount of amplicon produced (as measured by the peak height in relative fluorescence units - RFU). This is affected by the degradation status of the DNA and potential PCR inhibitors in the extraction eluent. Table 2 shows the peak height ratios calculated by comparing the heterozygote peak heights of the smaller D3S1358 amplicons to the larger Penta_E amplicons. Both blood and buccal samples yielded median ratios not exceeding 1, indicating limited skewing of the amplified products in the majority of the samples. Obvious skewing did however occur in three blood and three buccal samples (10% of samples) where peak height ratios were above 1.5, indicating problems in the amplification of the larger loci. This is only a potential issue when skewed amplification leads to peak RFU values below the stochastic and analytical thresholds. In our laboratory setting, this affected the results from one blood and one buccal sample (3% of samples), leading to the production of potential partial profiles, which may be disputed in a forensic setting. Representative electropherograms showing skewed amplification can be seen in Figure 2. Ease-of-use
Figure 2: Representative electropherograms indicating poor amplification of DNA samples. Capillary electrophoresis output showing all amplified STR alleles for DNA extracted from three study samples. A) STR profile obtained from an uninhibited, non-degraded sample showing no skewing of the amplified products. STR profiles obtained from inhibited or degraded DNA samples showing significant skewing in a B) buccal and C) blood sample.
Table 3 displays the various factors that were assessed when determining the usability of the collection methods in the post-mortem setting. While the collection of buccal swabs took approximately five times longer to perform than that of blood samples, no drying time was required before placing the buccal samples into the storage containers. While some difficulty was observed in obtaining buccal cell samples from eight individuals (27%), due to limited access to the oral cavity (rigor mortis and hospital-based intubation), 50% of the blood samples were difficult to obtain. The major challenges observed in the collection of blood samples were the presence of thickened blood and extreme clotting in some cases which poses a problem with transfer to and drying of the FTA card.
Discussion
A reliable method for obtaining DNA samples is essential to facilitate the efficient generation of STR profiles and successful identification of individuals in forensic investigations, including the identification of deceased individuals. In South Africa, as part of the new legislation, reference DNA samples from individuals will be collected using the EasiCollect device, which is used to obtain buccal epithelial cells from inside the mouth. The purpose of the present study was to evaluate the performance of this device in the mortuary setting, by comparing it to the current methodology of femoral blood on FTA cards.An estimation of the quantity of DNA present in a single 3 mm FTA punch showed that the median yield of total DNA for blood samples (5.54 ng) was significantly lower than that for buccal samples collected using the EasiCollect device (29.99 ng). Buccal DNA samples are reportedly often contaminated with bacteria DNA [11], a factor which may have contributed to the higher quantity of DNA measured in buccal samples in this study. However, qPCR of human DNA performed on a subset of the buccal samples in our study revealed very little non-human DNA contamination. Alternatively, the lower DNA yield average in blood samples could be due to the considerable number of samples which contained thickened or extremely coagulated blood (40%). Although blood clots have been found to be rich in DNA [25] due to their high white blood cell content, the poor absorption of these samples onto the FTA cards, as observed in this study, may result in a decrease in nucleic acid capture on the card matrix and thus result in lower yields. DNA quality was assessed using 260/280 nm absorbance ratios. Both sample types produced DNA contaminated with protein, with ratios well below the acceptable 1.8 ratio which is used as a reference for optimal DNA quality [23]. The buccal cell DNA quality results are comparable to previous studies [11,26], but the blood DNA quality was poorer than expected [11,27].
All DNA samples used in our study generated sufficient DNA for a PCR input value of >1 ng, which allows for optimal STR profiling [3], allowing both sample types to be used as a source of DNA. As shown in our study, all thirty individuals generated concordant results between the blood and buccal cell samples, illustrating that the DNA obtained from both collection methods was of sufficient quantity and quality for STR profiling. The amplification efficiency, measured by the peak height ratio of small to large markers, indicated that the majority of samples in this study showed relatively little skewing of the amplified products, signifying limited DNA degradation and PCR inhibition (due to collagen [28], hematin [28,29] or bacterial proteins [30]), despite the poor quality DNA (poor 260/280 nm ratio). When laboratory thresholds were applied to the STR profiles, amplification skewing resulted in only 3% of the profiles producing potential partial profiles, which represented both a single blood and buccal sample.
A blood sample is currently the specimen of choice for DNA collection during post-mortem examinations. As most bodies are dissected during the autopsy, the femoral vessels become accessible and a blood sample can subsequently be easily obtained and transferred onto the FTA card. It becomes more labour intensive when bodies are not autopsied due to obvious cause of death. Inthese cases, a blood sample may be obtained by an incision into thefemoral vessels or by palpating the vessel and collecting the blood using a needle and syringe. Although blood samples are quick and easy to collect, coagulated samples (which occurred in 40% of cases in our study) impede sample collection, transfer to the FTA cards, drying time and DNA yields. Currently, improvised kits for collection of blood samples in the mortuary are supplied by the South African Police Service (SAPS) and prepared within the nonsterile environment of the mortuary by untrained personnel. These kits not only lack tracking information necessary to ensure that the correct chain-of-custody of evidence is maintained, a vital aspect for criminal proceedings, but there is also a major DNA contamination issue, which is compounded by the need to dry the FTA cards in a busy state mortuary environment. The above mentioned factors limit the efficacy of blood samples stored on FTA cards as a suitable method for collecting DNA samples within the mortuary. In contrast, the EasiCollect device was easy to use and rigor mortis appeared not to prevent sample collection. The provision of the collection device in a convenient sterile packaging with tracking information and the ability to immediately store the sample without drying counters the problems experienced with the current blood collection techniques employed.
Several exclusions were made during the sampling process, due to prior investigation. One important exclusion from our study were infants younger than six months, as it was found that it was not possible to use the EasiCollect device due to the small size of their mouths. This will potentially be a significant problem when required to collect DNA samples from the countless concealed birth cases examined in mortuaries in South Africa. Unfortunately neither sample type could be obtained from badly decomposed or charred bodies. In such cases, alternative biological samples such as muscle [22], liver [2], brain tissue [31] or a fragment of the femoral head [32,33] would need to be obtained as a DNA sample. Additionally, further research will need to be conducted in order to determine if the EasiCollect device will be effective in collecting buccal DNA samples from individuals with extraneous substances in their oral cavities.
This study has demonstrated that both femoral blood transferred to FTA cards and buccal samples obtained by the EasiCollect device can be used to collect samples from deceased individuals in a state mortuary environment, which can then be used for DNA STR profiling. Due to the numerous issues associated with the collection of blood samples, this method of DNA collection is not optimal for use in the forensic setting. The greater yields derived from buccal samples, as well as the limited challenges associated with collecting samples makes the EasiCollect device a robust method for use within the mortuary, which we would recommend. It is important to acknowledge however, that one single method will not suffice for the collection of DNA samples within the mortuary. Complex situations may arise in which alternative biological specimens are required; therefore a variety of kits should be available for use.
Acknowledgements
The authors would like to acknowledge Marelette de Beer (Unistel Medical Laboratories, Cape Town, South Africa) for providing technical assistance.This project was supported by funding from the Division of Forensic Medicine, Department of Clinical Laboratory Services, University of Cape Town; National Research Foundation; as well as the Postgraduate Publication Incentive from the University of Cape Town Faculty Research Committee Awards. The funding sources had no involvement in the study design; in the collection, analysis and interpretation of data; or the writing of the report.
References
- Ghatak S, Muthukumaran RB, Nachimuthu SK (2013) A simple method of genomic DNA extraction from human samples for PCR-RFLP analysis. J Biomol Tech 24: 224-231.
- Hoff-Olsen P, Mevåg B, Staalstrøm E, Hovde B, Egeland T, et al. (1999) Extraction of DNA from decomposed human tissue. An evaluation of five extraction methods for short tandem repeat typing. Forensic Sci Int 105: 171-183.
- Norrgard K (2008) Forensics, DNA Fingerprinting, and CODIS. Nat Educ 1: 35.
- Myers BA, King JL, Budowle B (2012) Evaluation and comparative analysis of direct amplification of STRs using PowerPlex® 18D and Identifiler® Direct systems. Forensic Sci Int Genet 6: 640-645.
- INTERPOL (2013) Global DNA profiling survey results; 2012. Distributed at the 7th International DNA Users’ Conference for Investigative Officers, INTERPOL General Secretariat, Lyon, France.
- (2013) Act No. 37 of 2013: Criminal law (Forensic procedures) Amendment Act. South Africa.
- Mas S, Crescenti A, Gassó P, Vidal-Taboada JM, Lafuente A (2007) DNA cards: determinants of DNA yield and quality in collecting genetic samples for pharmacogenetic studies. Basic Clin Pharmacol Toxicol 101: 132-137.
- Cozier YC, Palmer JR, Rosenberg L (2004) Comparison of methods for collection of DNA samples by mail in the Black Women's Health Study. Ann Epidemiol 14: 117-122.
- Beckett SM1, Laughton SJ, Pozza LD, McCowage GB, Marshall G, et al. (2008) Buccal swabs and treated cards: methodological considerations for molecular epidemiologic studies examining pediatric populations. Am J Epidemiol 167: 1260-1267.
- Lindstrom JD (2012) A more effective means to collect & process reference DNA samples. U.S. Department of Justice.
- Hansen TV, Simonsen MK, Nielsen FC, Hundrup YA (2007) Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: comparison of the response rate and quality of genomic DNA. Cancer Epidemiol Biomarkers Prev 6: 2072-2076.
- Burger MF, Song EY, Schumm JW (2005) Buccal DNA samples for DNA typing: new collection and processing methods. Biotechniques 39: 257-261.
- Moore L, Wiencke J, Eng C, Zheng S, Smith A (2001) Evaluation of buccal cell collection protocols for genetic susceptibility studies. Biomarkers 6: 448-454.
- García-Closas M, Egan KM, Abruzzo J, Newcomb PA, Titus-Ernstoff L, et al. (2001) Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol Biomarkers Prev 10: 687-696.
- Milne E, van Bockxmeer FM, Robertson L, Brisbane JM, Ashton LJ, et al. (2006) Buccal DNA collection: comparison of buccal swabs with FTA cards. Cancer Epidemiol Biomarkers Prev 15: 816-819.
- Klassen TL, von Rüden E, Drabek J, Noebels JL, Goldman AM (2012) Comparative analytical utility of DNA derived from alternative human specimens for molecular autopsy and diagnostics. J Mol Diagn 14: 451-457.
- Mulot C, Stücker I, Clavel J, Beaune P, Loriot MA (2005) Collection of human genomic DNA from buccal cells for genetics studies: comparison between cytobrush, mouthwash, and treated card. J Biomed Biotechnol 3: 291-296.
- GE Healthcare Life Sciences (2011) STR amplification of DNA from buccal and blood bamples btored long-term on WhatmanTM FTATM Cards.
- Fujii K, Kitayama T, Nakahara H, Mizuno N, Yoshida K, et al. (2009) Comparison of buccal cell collection kits for STR typing. Jpn J Forensic Sci Tech 14: 55-60.
- Wieczorek D, Krenke B (2009) Direct amplification from buccal and blood samples preserved on cards using the PowerPlex® 16 HS System.
- (2014) Government gazette No 38074: Notice calling for Public Comments; Criminal Law (Forensic Procedures) Amendment Act, Act 37 of 2013: Forensic DNA Regulations. The Government Printer, Pretoria.
- Hansen J, Lesnikova I, Funder AM, Banner J (2014) DNA and RNA analysis of blood and muscle from bodies with variable postmortem intervals. Forensic Sci Med Pathol 10: 322-328.
- Thermo Scientific (2013) Assessment of nucleic acid purity. TO42-Technical Bulletin. Thermo Fisher Scientific.
- Wang DY, Chang CW, Oldroyd NJ, Hennessy LK (2009) Direct amplification of STRs from blood or buccal cell samples. Forensic Sci Int 2: 113-114.
- Bär W, Kratzer A, Mächler M, Schmid W (1988) Postmortem stability of DNA. Forensic Sci Int 39: 59-70.
- Rogers NL, Cole SA, Lan HC, Crossa A, Demerath EW (2007) New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am J Hum Biol 19: 319-326.
- Song Y, Fahs A, Feldman C, Shah S, Gu Y, et al. (2013) A reliable and effective method of DNA isolation from old human blood paper cards. Springerplus 2: 616.
- McCord B, Opel K, Funes M, Zoppis S, Meadows Jantz L (2011) An investigation of the effect of DNA degradation and inhibition on PCR amplification of single source and mixed forensic samples.
- Perch-Nielsen IR, Bang DD, Poulsen CR, El-Ali J, Wolff A (2003) Removal of PCR inhibitors using dielectrophoresis as a selective filter in a microsystem. Lab Chip 3: 212-216.
- Wilson IG (1997) Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 63: 3741-3751.
- Motani H, Sakurada K, Akutsu T, Ikegaya H, Hayakawa M, et al. (2006) Usefulness of dura mater in providing DNA samples for identifying cadavers. J Forensic Sci 51: 888-892.
- Westen AA, Gerretsen RR, Maat GJ (2008) Femur, rib, and tooth sample collection for DNA analysis in disaster victim identification (DVI): a method to minimize contamination risk. Forensic Sci Med Pathol 4: 15-21.
- Mundorff A, Davoren JM (2014) Examination of DNA yield rates for different skeletal elements at increasing post mortem intervals. Forensic Sci Int Genet 8: 55-63.