Journal of Emergency Medicine & Critical Care
Coagulase-Negative Staphylococci Isolated from Blood Cultures: Contamination or Infection? Atherapeutic and Diagnostic challenge
Distasi MA* Tarricone NR and Del Gaudio Tito
- U.O.C. di Patologia Clinica P.O. di Andria, ASL BT, Italy
*Address for Correspondence: Distasi Maria Antonietta, U.O.C. di Patologia Clinica P.O. di Andria, ASL BT viale Istria, 176123 Andria (BT) Italy, E-mail: mailto:mariadistasi.md@libero.it
Citation: Distasi MA, Tarricone NR, Tito DG. Coagulase-Negative Staphylococci Isolated from Blood Cultures: Contamination or Infection? A therapeutic and Diagnostic challenge. 2017;3(1): 3.
Copyright: © 2016 Dikme O. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Emergency Medicine & Critical Care | ISSN: 2469-4045 | Volume: 2, Issue: 1
Submission: 30 January, 2017| Accepted: 22 February, 2017 | Published: 06 March, 2017
Abstract
The possibility of contamination of the blood cultures, particularly by Coagulase-Negative Staphilococci (CoNS), is one of the challenges in microbiology. We performed a study from 2010 to 2015 to evaluate the percentage of CoNS arising from probable contamination and the percentage of CoNS probably responsible for sepsis. Additionally, we analysed susceptibility to antibiotics of all found microorganisms. In the observation period, we performed 18065 blood cultures with blood drawn from 4199 patients (averaging 4.3 blood cultures per patient). We used Tokar’s algorithm to layer positive blood cultures. Identification and antibiogram were performed with the Vitek2 Compact system (BioMèrieux). From 2010 to 2015 we found 1558 positive patients: bacteremia was considered very probable in 279 patients (6.6% of total patients, 17.9% of positive samples), bacteremia was considered as arising from contaminants in 510 patients (12.1% of total patients, 32.7% of positive samples), bacteremia was considered undetermined in 12 patients (0.3% of total patients, 0.8% of positive samples). Out of all the found strains, sensitivity to antibiotics was as follows: Penicillin, 3.6%; Oxacillin, 15.1%; Eritromicin, 22.3%; Vancomicin, 100%; Daptomicin, 98.5; Teicoplanin (except S. epidermidis and S. haemolyticus), 94.9%. In our laboratory, we execute identification and antibiogram also in the case of probable contamination. In this case, the positive bottle is reported with a note that defers the treatment decision to the clinician. The analysis of the SIR index showed that CoNS can represent a therapeutic challenge because CoNS have high methicillin resistance (84.9%). On the other hand, sensitivity to Vancomicin, Daptomicin, and Teicoplanin offers the possibility of an alternative therapeutic route.
Keywords
Blood culture; CoNS; Staphylococcus; Bacterial contamination; Tokars; Teicoplanin
Introduction
Blood culture is the most appropriate tool for the diagnosis of the cause of sepsis. The diagnostic efficacy of blood cultures are closely related to technical detailsof the execution of the blood draw and the number of samplings. One of the diagnostic challenges is represented by the possibility of contamination of blood cultures, in particular by Coagulase-negative staphylococci (CoNS) [1]. We performed a study from 2010 to 2015 to evaluate the percentage of CoNS arising from probable contamination and the percentage of CoNS that is probably responsible for sepsis. We finally provide an analysis of susceptibility to antibiotics.
Material and Methods
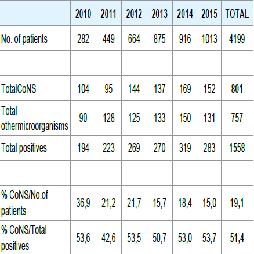
During the observation period, 18065 blood cultures taken from 4199 patients (mean 4.3 blood cultures / patient) Table 1 were submitted to our laboratory. We asked to perform at least three sets of blood draws, each consisting of a bottle for aerobic microorganisms and one for anaerobic microorganisms, after thorough disinfection of the patient's skin and the operator's hands [2]. Blood cultures were performed using the BacT/Alert FA bottles for aerobic microorganisms, BacT/Alert FN for anaerobic microorganisms, BacT/Alert PF for pediatric patients, all incubated for 7 days in the BacT/Alert 3D system (bioMerieux). In the case of positivity, the blood cultures were subsequently respectively seeded on Enriched Chocolate Agar incubated for 24 h at 36 °C in CO2, Mac Conkey Agar, Mannitol Salt Agar, MRSA-selective Agar, and Sabouraud Agar incubated for 24 h at 36 °C under aerobic conditions. In addition to the above culture media, the samples positive for anaerobic microorganisms were inoculated on Enriched Chocolate Agar, Columbia CNA Agar, and incubated for 24 h at 36 °C under anaerobic conditions. Identification tests and susceptibility to antibiotics tests were performed by Vitek 2 Compact system (bioMerieux). We executed antibiograms using the following cards: from January 2010 to February 2012, AST-P580; from March 2012 to April 2013, AST-P619; from May 2013 to December 2015, AST-P632. For the stratification in positive blood cultures of infectious agents, contaminants, and undetermined material, we referred to Tokar’s algorithm: very probable bacteremiais assigned to samples tested positive for two or more bottles per battery; improbable bacteremia was assigned to samples tested positive for one bottle per battery; finally, samples that tested positive only on one blood culture were considered undetermined [3].
Results
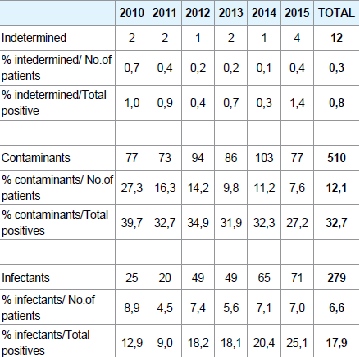
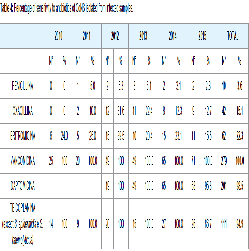
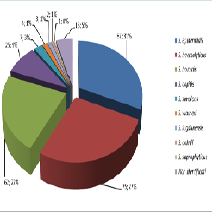
From the year 2010 to 2015 1558 patients were found positive, of which 801 for CoNS (19.1% of patients; 51.4% of the positive samples) Table 2. Of the patients that were found positive, 279 patients (6.6% of patients; 17.9% of the positive samples) were associated to very likely bacteremia, 510 (12.1% of patients and 32.7% of the positive samples) were associated to probable contaminants, and 12 patients (0.3% of patients; 0.8% of the positive samples) were indeterminate Table 3. From samples associated to very likely bacteremia, we isolated S. epidermidis in 87 samples (31.2%), S. haemolyticus in 75 samples (26.9%), S. hominis in 62 samples (22.2%), S. capitis in 25 samples (9.0%), S. simulans in 7 samples (2.5%), S. warneri in 4 samples (1.4%), S. lugdunensis in 3 samples (1.1%), S. cohnii in 2 samples (0.7%), S. saprophyticus in 1 sample (0.4%) Figure 1. For 13 strains (4.7%), identification was not possible. On all isolates, sensitivity to antibiotics was determined as follows: 3.6% for Penicillin, 15.1% for Oxacillin, 22.3% for Erythromycin, 100% for Vancomycin, 98.5% for Daptomycin, 94.9% for Teicoplanin (except for S. epidermidis and S. haemolyticus) Table 4.
Discussion
In our observation period, we verified, in agreement with the literature data, that increasing the number of patient samples improves the likelihood of identifying likely sepsis by CoNS. Additionally, an increased number of samples allows for an easier layering of those CoNS that are to be considered likely contaminants [4]. In our laboratory, even in the case of probable contamination, we none the less perform identification and an antibiogram. A positive bottle is reported along with negative bottles in the same battery with a note (Presence of organism due to probable contamination. Please check the disinfection process of the skin, then clinically assess the need for antibiotics) which has the goal of signaling the probable contamination and to delegate to the clinician any treatment decisions given the clinical condition of the patient. A significant problem was posed by the samples treated as indeterminate, which should not be interpreted as having any diagnostic significance. In fact, in our study, the 12 indeterminate samples were taken from pediatric patients, in whom the difficulty of execution of the blood draw hinders the possibility of obtaining more than one sample. In the case of indeterminate samples, the identification of CoNS is still performed and the relative susceptibility is reported to the clinician, who is to assess whether and how to treat the patient. The analysis of antibiotic susceptibility showed, and in agreement with the literature data [5,6] 84.9% resistance to Methycillin. In the case of Teicoplanin, in particular with reference to S. epidermidis and S. haemolyticus, the system that we have used for sensitivity testing does not deem the result of resistance to the drug significant [7]. Furthermore, we observed, in the course of 2015, a reduction of sensitivity of CoNS to Erythromycin and the appearance of the first strains resistant to Teicoplanin and Daptomycin, probably due to the intense use of these antibiotics in our hospital. We must point out that the use of the Tokar's algorithm is a limit of our work: this method is not accepted as a standard for the stratification of contaminating micro-organisms; however, it seemed the most appropriate to the characteristics of our retrospective study: our aim was to suggest a correction of the levy implementation procedures for blood culture above all about the number of sets to be sent to the laboratory. Finally, being a retrospective study has been impossible to verify whether our observations regarding the prediction of infection or contamination coincided with clinical evidence of these and what was the eventual therapeutic choice.
Conclusion
CoNS can represent for the clinician a serious therapeutic challenge, especially because it is characterized by methicillin resistance. However, the high percentage of Vancomicin, Daptomycin and Teicoplanin sensitivity can offer an adequate alternative therapeutic choice. Blood cultures positive for CoNS pose a challenge both for the laboratory, and for the clinician. Closer cooperation between the two sectors is essential, on the one hand, to improve laboratory performance, with respect to the adequacy of the results and sample handling costs; on the other hand, it will lead to better clinical therapeutic choices for the patient, including the decision to not engage in any therapy. This latter decision could be effective to prevent the onset of resistance to other drugs.
References
- Camporese A (2007) Numero di emocolture solitarie e percentuali di contaminazione come indicatori della qualità preanalitica delle emocolture. RIMeL - IJLaM 3: 106-112.
- Gruppo di Lavoro Infezioni Paziente Critico (GLIPaC) (2014) Infezioni del torrente circolatorio. Proposta di Percorso Diagnostico presentato durante in XXXVII congresso Nazionale AMCLI.
- Tokars JI (2004) Predictive Value of blood cultures positive for coagulase negative Staphylococci: implications for patient care and health care quality assurance. Clin Infect Dis 39: 333-341.4.
- Conti A, De Rosa R (2008) Quality assurance della emocoltura. RIMeL - IJLaM 4: 36-44.
- Asaad AM, Ansar Qureshi M, Mujeeb Hasan S (2016) Clinical significance of coagulase-negative staphylococci isolates from nosocomial bloodstream infections. Infect Dis (Lond) 48: 356-360.
- Obolski U, Alon D, Hadany L, Stein GY (2014) Resistance profiles of coagulase-negative staphylococci contaminating blood cultures predict pathogen resistance and patient mortality. J Antimicrob Chemother 69: 2541-2546.
- VITEK® 2 Systems Software 05.03 Addendum Customer Communication