Journal of Clinical and Investigative Dermatology
Download PDF
Research Article
NOTCH1 in Cutaneous Squamous Cell Carcinoma Arising in Immunosuppressed Patients: A Systematic Review and Quantitative Analysis
Miller AD*, Chow ML and Brian Jiang SI
Department of Dermatology, University of California San Diego, USA
*Address for Correspondence: Miller AD, Department Of Dermatology, University Of California San Diego,8899 University Center Ln St. 350, San Diego, CA, 92122; USA; Tel: 515-
573-0935; Email: Adm005@Health.Ucsd.Edu
Submission: 11 August, 2021;
Accepted: 22 November, 2021;
Published: 25 November, 2021
Copyright: © 2021 Miller AD, et al. This is an open access article distributed
under the Creative Commons Attribution License, which permits unrestricted
use, distribution, and reproduction in any medium, provided the original work
is properly cited.
Abstract
Immunosuppression is a strong risk factor for cutaneous squamous
cell carcinoma (cSCC). Immunosuppression is also associated with
unique mutagenic stressors that likely contribute to cSCC pathogenesis.
However, it is unknown whether these stressors contribute to a distinct
mutation profile that may drive disease progression. This review was
conducted to assess the mutational landscape of cSCC arising in
immunosuppressed hosts. Specifically, we sought to determine gene
mutation frequencies in immunosuppressed cSCC. An electronic
search was performed in PubMed, Embase, Scopus, and Cochrane
databases. Studies performing DNA sequencing or genotyping of
cSCC were identified. Studies were excluded if the immune status
of each tumor was not available. Eighteen studies met inclusion
criteria. Due to study heterogeneity a meta-analysis was unable to be
performed. However, statistical analysis was performed on the most
frequently reported genes. NOTCH1 was the most frequently mutated
gene in immunosuppressed cSCC, and was significantly higher than
immunocompetent cSCC after multiple comparison adjustment (77.7
versus 58.1%, OR 2.50, 95% CI 1.40-4.46, p=0.002). No other statistically
significant differences were observed. Our results suggest that NOTCH1
mutations are more common in cSCC arising in immunosuppressed
hosts. Several prior observations reviewed here further support a role
for NOTCH1 in immunosuppressed cSCC, however larger studies are
needed to confirm our findings.
Introduction
Cutaneous squamous cell carcinoma (cSCC) is the second most
common malignancy in Caucasians with an increasing incidence
worldwide [1]. While surgery is curative in most cases, locally
advanced and metastatic disease is associated with significant
morbidity and mortality [2]. High-risk clinical and histologic features
correlate with prognosis and have been incorporated into staging
criteria [3]. However, the genetic predictors of advanced disease
remain poorly understood.
These genetic predictors are difficult to investigate as a result
of the number and types of mutations present. At 50 mutations per
megabase pair of coding DNA, the tumor mutation burden in cSCC is
significantly higher than any solid organ or hematologic malignancy
[4,5]. Mutations affect diverse pathways involving keratinocyte
differentiation, cell-cycle regulation, cellular proliferation, and
chromatin maintenance. Additionally, histologically normal skin
from sun-exposed areas has a mutation rate equal to most human
cancers, making identification of cSCC driver mutations particularly
difficult [6]. Gross chromosomal aberrations and widespread
epigenetic dysfunction due to DNA methylation, mutations in
noncoding DNA, and variable expression of noncoding RNA molecules further complicate the genomic landscape of cSCC [7-10].
An important risk factor for cSCC is immunosuppression, with
the highest risk observed in organ transplant recipients (OTR). In
OTR, the risk of developing cSCC is 65 to 100-fold higher than the
general population [11]. Cutaneous SCC in OTR also have greater
propensity for aggressive subclinical extension, local recurrence,
metastasis, and death [12-15]. The clinical and histologic features
predictive of poor outcome in immunosuppressed (IS) cSCC are
similar to those in immunocompetent (IC) cSCC [1]. Importantly,
immunosuppression is associated with unique mutagenic stressors
that likely contribute to genetic instability leading to cutaneous
oncogenesis. Despite these unique stressors, few studies have directly
compared the mutation profiles of IS and IC cSCC.
Therefore, the purpose of this review is to provide a quantitative
summary of mutation frequencies in IS and IC cSCC. By doing so, we
hope to aid in the discovery of differentially mutated genes that may
contribute to the more aggressive phenotype observed in IS hosts.
Methods
Search Strategy and Study Selection:
A systematic search of PubMed, Embase, Scopus, and Cochrane
databases was completed by two authors (ADM, MLC) from each
database’s earliest inception to April, 2020. Search terms included
“cutaneous squamous cell carcinoma”, “genetics”, and “mutation”.
Bibliographies of articles were reviewed for additional relevant
studies. Studies were initially screened by article title and abstract.
Studies deemed relevant based on screening criteria were reviewed in
full to establish a final set of studies.Inclusion and Exclusion Criteria:
Original studies in which DNA sequencing or genotyping of
cSCC was performed were included in the analysis. Upon screening,
studies were excluded for any of the following: (1) studies consisting
exclusively of actinic keratoses, squamous cell carcinoma in situ,
keratoacanthoma, and/or non-cutaneous SCC; (2) studies in subjects
with predisposing genetic conditions; (3) studies of cSCC arising
secondary to treatments based on BRAF-inhibition, psoralen and
ultraviolet A, radiation, or arising in chronic wounds; (4) studies utilizing human cell lines; (5) studies with indiscernible immune
status; and (6) studies utilizing techniques other than DNA sequencing,
small nucleotide polymorphism (SNP) microarray, or microsatellite
analysis (e.g., single strand conformational polymorphism analysis).Data Collection, Quality Assessment, and Risk of Bias:
The mutation status of each tumor was recorded as a binary
outcome (mutated/wild-type) for the genes reported in four or more
studies. Limitations of each study were sought and disclosed. The
limitations affecting study quality and contributing to potential bias
include: (1) small sample sizes, (2) unequal group sizes, (3) varying
definitions of immunosuppression, and (4) varying methods and
assays used for mutation analysis.Statistical Analysis
We initially sought to perform a meta-analysis. However, there
was considerable interstudy variability in terms of methods for gene
analysis and how immunosuppression was defined. Additionally,
four studies included either IS or IC samples, but not both. Therefore,
appropriate statistical analysis using a random effects model was
not possible. Instead, data from studies were combined to calculate
a single mutation frequency and odds ratio for each gene reported
in at least six separate studies. Fisher’s exact test was used to assess
statistical significance between the two groups,and an unweighted
odds ratio was used to estimate effect size. Multiple comparisons were
adjusted using the Bonferroni correction with statistical significance
set at p < 0.003 (type I error rate, α, of 0.05 with 17 separate gene
comparisons).
Results
Search Results:
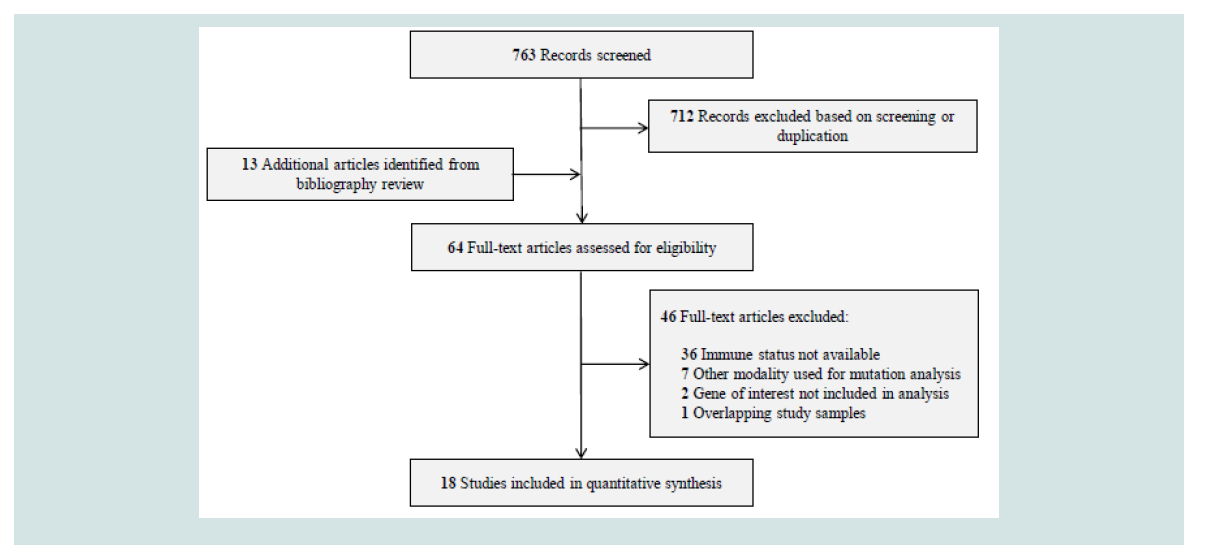
A flowchart of our selection process is depicted in Figure 1. A total
of 763 articles resulted from our literature search and 13 articles were
identified through bibliography review. After screening by title and abstract, 712 studies were excluded due to ineligibility or duplication. The remaining 64 studies were reviewed in full text, and 18 studies
were ultimately included in our analysis [4,17-33].Studies and Genes:
The study characteristics are shown in Supplementary Table 1.
The number of genes analyzed in each study ranged from a single
gene to whole exome analysis. DNA sequencing was performed in
all studies, and the types of mutations detected varied depending
on the methods used. Additionally, genotyping was performed in
four studies using SNP or microsatellite analysis to detect genespecific
loss-of-heterozygosity (LOH), copy number alterations, and
microdeletions. The 18 studies include a total of 601 cSCC: 264 from
IS and 337 from IC subjects. Mutation status was recorded for the 136
genes analyzed in at least four studies. The number of cSCC for which
mutation status was available varied depending on the genes included
in each study: the range was 37-156 tumors per gene for IS and 69-232
tumors per gene for IC cSCC.Quantitative and Statistical Analysis:
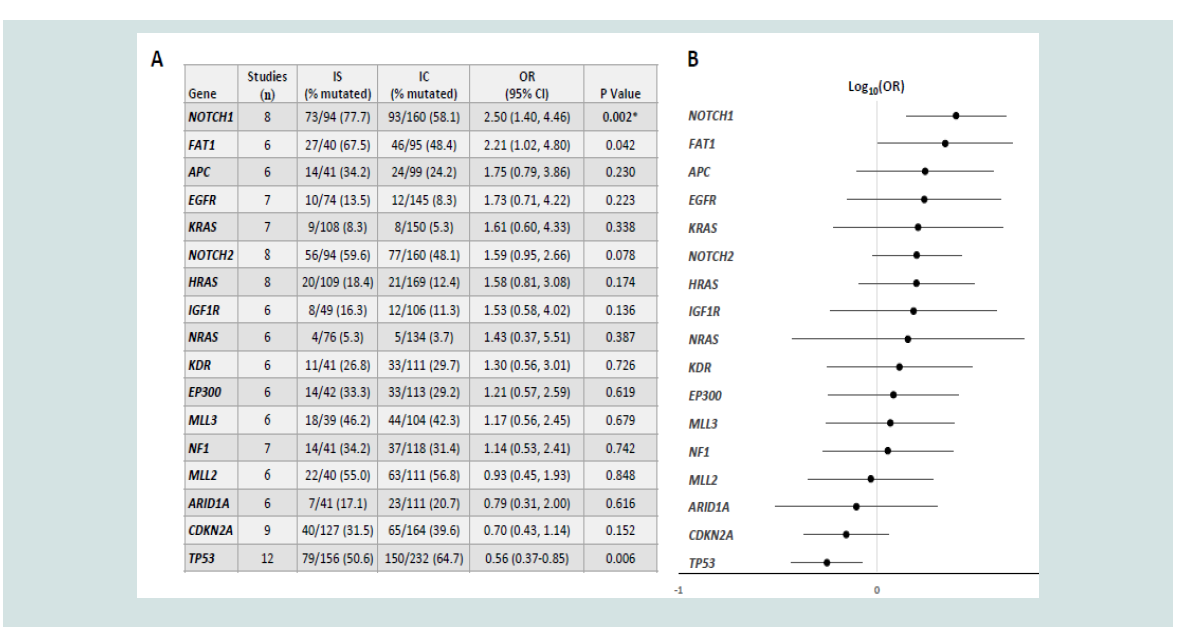
The mutation frequencies of the 136 genes reported in four or
more studies are shown in Supplementary Table 2 The mutation
frequencies with corresponding p-values and odds ratios for the 17
genes reported in six or more studies are shown in Figure 2. NOTCH1
was the most frequently mutated gene in IS cSCC (77.7%) and overall
(65.4%). Additionally, the frequency of NOTCH1 mutations was
significantly higher in IS versus IC cSCC (77.7 versus 58.1%, OR 2.50,
95% CI 1.40-4.46, p=0.002).TP53 was more frequently mutated in IC
versus IS cSCC (64.7 versus 50.6%), however this was not statistically
significant after multiple comparison adjustment (p=0.006).No other
statistically significant differences were observed.Discussion
This study aimed to review the literature and calculate gene mutation frequencies in IS cSCC in order to better understand its genetic determinants. Despite our efforts, a meta-analysis could not
be performed due to the scarcity and heterogeneity of existing data.
However, our review and analysis suggest that NOTCH1 may be
preferentially mutated in IS cSCC.
Ultraviolet (UV) radiation, particularly UVB, is the most
important risk factor forall non melanoma and melanoma skin
cancers. This is demonstrated by the predominance of C->T
mutations, which are the hallmark UVB-induced mutagenesis[4].In
addition to chronic UVB exposure, immunosuppression is associated
with unique mutagenic stressors that may contribute to cSCC
oncogenesis through distinct genetic mechanisms. These stressors
may act synergistically with UV light or through UV-independent
mechanism. This is supported by fewer UVB-associated mutations
observed in IS cSCC [34]. Additionally, clinical evidence supporting
a UV-independent mechanism is demonstrated by the observation
that cSCC in OTR may predict development of subsequent noncutaneous
SCC, particularly of the oropharynx and lung, suggesting
an internal carcinogenic driver [35].Through UV-dependent
mechanisms, both calcineurin inhibitors and mycophenolate mofetil
inhibit nucleotide excision repair enzymes leading to persistence of
UVB-induced cyclopyrimidine dimers and reduced cellular apoptosis
[36,37]. Conversely, the purine analog azathioprine sensitizes cells to
UVA-mediated oxidative DNA damage. This mechanism is distinct
from the UVB-induced mutations, as demonstrated by the unique
cSCC mutation signature observed in patients receiving azathioprine
[22]. Separate from the mutagenic effects of immunosuppressive
medications, unique mutation profiles have been observed in head
and neck SCC (HNSCC) associated with HIV and HPV infection
[38,39]. These mutations may be due to insertional mutagenic events or a host defense mechanism meant to protect against retroviral
infection, respectively.
Despite the unique and numerous mutagenic stressors related to
immunosuppression, the overall mutation burden does not appear to
be higher in IS compared to IC cSCC. While one early microsatellite
analysis study observed the rate of LOH in OTR to be less than half
of that in IC cSCC [40], a subsequent study using higher resolution,
genome-wide SNP microarray found no difference in rate of LOH
between IS and IC cSCC[41]. Instead, they demonstrated the number
of chromosomal aberrations correlated with the degree of tumor
differentiation, a finding that has been reproduced [22]. Similarly,
targeted gene and whole-genome sequencing studies found no
difference in overall mutation burden based on immune status [9,34].
To date, few studies have compared the specific genetic alterations
of IS and IC cSCC. In a targeted sequencing study, no difference in
mutation frequency of seven driver genes in 52 IS and 39 IC cSCC [4].
Similarly, a whole-exome sequencing study found no difference in 22
significantly mutated genes in 33 IS and 7 IC cSCC [22]. These studies
suggest that cSCC share common driver mutations regardless of the
underlying immune status of the host. An earlier study performed by
Ridd et al., sought to characterize gene mutation status and protein
expression of six receptor tyrosine kinases known to be mutated in a
subset of cSCC [31]. They found that EGFR protein over expression
was significantly higher in non-OTR compared to OTR. However,
mutations and amplifications of the EGFR gene were exceedingly
rare in both groups, suggesting posttranscriptional modifications
contributing to protein over expression. Similar discordance between
EGFR protein over expression and gene amplifications was reported
by Cañueto et al., however no difference in protein over expression
was observed this study based on immune status [42]. Mutations in the tumor suppressor CDKN2A have also been studied with conflicting
results. Brown et al.,observed CDKN2A alterations more frequently
in IC cSCC[19], while Mühleisen et al., demonstrated reduced allelic
imbalance at chromosome 9p21 containing CDKN2A [43]. Clearly,
a knowledge gap exists in regards to the specific genetic alterations
occurring in IS cSCC due to limited studies and conflicting data
Our quantitative analysis suggests that mutations in NOTCH1 are
more common in IS cSCC. The NOTCH genes encode transmembrane
receptors with tissue specific function [44].In the skin NOTCH1
promotes terminal differentiation of keratinocytes, and several lines
of evidence demonstrate its role as a tumor suppressor in squamous
epithelium [44-46].In addition to our quantitative analysis, several
prior observations implicate the NOTCH pathway specifically in IS
cSCC.
First, there is a complex relationship between NOTCH and human
papillomavirus (HPV).Notably, β-HPV E6 protein directly inhibits
the primary cofactor of NOTCH1, MAML1, resulting in decreased
expression of its target genes [47]. Similarly, E6 protein inhibits
transcription of NOTCHvia p53 inhibition [48]. A transposonmediated
insertional mutagenesis protocol in mice demonstrated that
HPV infection decreased the threshold of NOTCH1 loss necessary
for oral SCC carcinogenesis [49]. A similar sensitizing effect may exist
in cSCC arising in IS patients co-infected with HPV. Although this
may predict a lower mutation frequency ofNOTCH1in IS cSCC, this
assumption is an oversimplification. Specifically, NOTCH1 can play
a dual role as either a tumor suppressor or oncogene in squamous
epithelium depending on the overall mutational context and the
presence of HPV infection [49]. In addition to HPV, calcineurin
inhibitors likely contribute to IS cSCC carcinogenesis through a
NOTCH-dependent mechanism. Specifically, NOTCH functions
upstream of calcineurin/NFAT in an integrated pathway promoting
keratinocyte terminal differentiation [50]. Lastly, it is possible that
unique mutagenic stressors in IS cSCC preferentially alter regions
within the NOTCH1 locus. When considering our results in the
context of the above findings, there is compelling evidence supporting
a role for NOTCH1 alteration in IS cSCC.
In addition to NOTCH1in IS cSCC, several other genetic
domains are primed for future study. Perhaps the most intriguing are
epigenetic alterations and TERT. In OTR, germline polymorphisms
in MTHFR confer an increased risk for cSCC [51]. Additionally,
OTR harboring these MTHFR polymorphisms were found to have
higher global levels of DNA methylation in both cSCC and unaffected
skin, suggesting an inherited risk due to aberrant DNA methylation
and epigenetic dysfunction [52]. While our quantitative analysis did
not detect differential mutation frequencies of genes involved in
chromatin remodeling and repair, examination of other epigenetic
determinants, including methylation signatures and noncoding RNA
expression, may provide valuable insight. Additionally, mutations
in the TERT promoter (TERTp) are present in 32% of IC cSCC and
associated with poor outcome [53]. Interestingly, Perrem et al., found
that telomeres in cSCC arising in OTR were significantly longer than
those arising in non-OTR [54]. Whether activating TERTp mutations
contribute to longer telomere length in IS cSCC warrants further
investigation, as the studies included in this review and quantitative
analysis were conducted on exonic DNA. Thus, the promoter
sequence was not analyzed.
Conclusion
Despite the growing understanding of the genetic landscape
of cSCC, the specific genetic determinants underlying IS cSCC
pathogenesis remain poorly understood. Further investigation into
this topic may help identify genetic drivers that could be targeted to
better prevent and treat cSCC arising in IS patients. We propose that
dysfunction in the NOTCH pathway, includingNOTCH1 mutations,
is of critical importance in the pathogenesis of cSCC arising in IS
patients and merits further investigation.