Journal of Analytical & Molecular Techniques
Download PDF
Research Article
*Address for Correspondence: Sylvain Ladame, Department of Bioengineering, Imperial College London, South Kensington Campus, London SW7 2AZ, U.K, Tel: (+44) 02075945308; E-mail: sladame@imperial.ac.uk
Citation: Meguellati K, Koripelly G, Ladame S. Single Nucleotide Polymorphism Detection Using a Biocompatible, Fluorogenic and DNA-Templated Reaction of Cyanine Dye Formation. J Analyt Molecul Tech 2013;1(1): 5.
Copyright © 2013 Ladame et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Analytical & Molecular Techniques |ISSN: 2474-1914 | Volume: 1, Issue: 1
Submission: 19 September 2013 | Accepted: 13 December 2013 | Published: 18 December 2013
DNase- and RNase-free water, potassium phosphate buffer and Calf-Thymus DNA were purchased from Sigma-Aldrich. PNA monomers were purchased from ASM Research Chemicals (Germany) and Rink amide resin for solid phase synthesis from Merck Biosciences (UK). DNA oligonucleotides were purchased from Sigma and were all HPLC purified (Table 1). Synthesis and Characterisation (HR-MALDI) of both purified fluorogenic PNAs (Pna1 and Pna2) was recently reported by us [31,32].
We first investigated the influence of the distance (i.e. number of nucleotides) between both PNAs on the efficiency of the fluorogenic reaction. Five DNA strands (WT0-4, Table 1), all containing both sequences complementary to Pna1 and Pna2 but separated by a variable number of nucleobases (from 0 to 4), were evaluated as potential candidates capable of templating the reaction of cyanine dye formation. While short distances between both PNAs (WT0-2) proved equally good at templating the fluorogenic reaction, slightly more flexible systems (e.g. WT3 where PNA/DNA duplexes formed upon hybridisation of Pna1 and Pna2 with the DNA template are separated by three nucleotides) appeared more favorable (Figure 3). This tendency was confirmed when increasing the distance even further (up to 7 nucleotides, data not shown), although never exceeding the maximum fluorescence observed with a 3 nucleotide gap (WT3).
Next, both WT2 and WT3 were selected for a more detailed study. The effect of systematic point mutations within the DNA template on the reaction efficiency was investigated. In the case of WT3, none of the single mutations tested led to a significant change in reaction efficiency, regardless of the nature and position of the mutation (Figure 4A). Significant inhibition of the fluorogenic reaction was observed only when mutating at least two consecutive residues in WT3 while near-complete inhibition was obtained when mutating a minimum of three residues, hence preventing hybridization of at least one of the fluorogenic PNAs (Figure 4B).Although WT3 seemed optimal for templating the reaction of cyanine dye formation, it appeared that a 3-nucleotide gap between both PNA complementary sequences was probably making the PNA-DNA complex too flexible to be responsive to minimal changes such as single mutations.
A significant decrease (up to 5-fold) in fluorescence intensity was observed when mutating one nucleotide within the sequence of WT2 complementary to Pna2 (Mut1-5, Table 1). As previosuly observed by others, the strongest “inhibitory” effects were obtained with single mutations located near the indole fluorogenic probe (e.g. strong>Mut1 vs Mut5). However, the nature of the mutation had no influence on the reaction time-course. For instance, similar time courses were obtained for Mut1, regardless of the nature of X (A,C or G). Surprisingly, DNA strands carrying mutations within the sequence of WT2 complementary to Pna1 (Mut6, Table 1) were found to be almost equally efficient templates when compared to the native sequence WT2. This is likely due to a greater conformational freedom of the Fischer’s base aldehyde allowing its reaction with the neighbouring indole even if Pna1 is not fully hybridized to the DNA template. Altogether, these results suggest that a constrained system in which two 5-mer fluorogenic PNAs can hybridize, simultaneously and two nucleotides apart, to a unique DNA strandserving as a reaction template, can be used for SNPs detection. Most sensitive detection is achieved when the mutation is located opposite the indole’s neighbouring nucleobase (Mut1) although the system remains sensitive enough for detecting mutations located further away in the DNA template (Mut2-4).
Single Nucleotide Polymorphism Detection Using a Biocompatible, Fluorogenic and DNA-Templated Reaction of Cyanine Dye Formation
Kamel Meguellati1, Girish Koripelly1 and Sylvain Ladame1,2*
- 1ISIS Universite de Strasbourg, 8 allée Gaspard Monge, BP 70028, 67083 Strasbourg, France
- 2Department of Bioengineering, Imperial College London, South Kensington Campus, London SW7 2AZ, U.K
*Address for Correspondence: Sylvain Ladame, Department of Bioengineering, Imperial College London, South Kensington Campus, London SW7 2AZ, U.K, Tel: (+44) 02075945308; E-mail: sladame@imperial.ac.uk
Citation: Meguellati K, Koripelly G, Ladame S. Single Nucleotide Polymorphism Detection Using a Biocompatible, Fluorogenic and DNA-Templated Reaction of Cyanine Dye Formation. J Analyt Molecul Tech 2013;1(1): 5.
Copyright © 2013 Ladame et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Analytical & Molecular Techniques |ISSN: 2474-1914 | Volume: 1, Issue: 1
Submission: 19 September 2013 | Accepted: 13 December 2013 | Published: 18 December 2013
Abstract
Water-soluble Peptide Nucleic Acids (PNAs) functionalised with fluorogenic cyanine dye precursors (i.e. a Fisher’s base aldehyde and a 2-methylene indolenine) are employed for detecting single-nucleotide polymorphism in vitro under near physiological conditions of salt and pH. This quick and versatile fluorogenic system has been designed so that fluorescence is formed only upon simultaneous hybridisation of both PNAs to a fully complementary DNA strand. The reaction yield increased up to around 10% upon addition of the DNA template, even in the presence of a large excess of Calf-Thymus competitor DNA.Introduction
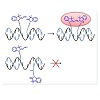
Since the final sequencing of the human genome was completed, in 2003, considerable research effort has been devoted to the identification and analysis of variations among individual genomes [1-4]. Although more than 99.9% of human DNA sequences are the same across the population, variations can have a major impact on how humans respond to diseases or drug therapies [5,6]. Amongst the most common forms of genomic variation, single-nucleotide polymorphism (SNP) accounts for over 80% of sequence variants [7-9]. Current estimates suggest that SNPs occur as frequently as every 100 to 300 bases and that they are stably inherited, making them a unique tool/marker for identifying numerous genetic and inherited complex diseases such as cancers and diabetes [7-9]. Therefore, there is an increasing need for high throughput technologies suitable for rapid and sensitive SNP genotyping [10-12]. So far, methods using short oligonucleotides or oligonucleotide mimics as probes to reveal the presence of complementary sequences have been successfully developed [13,14]. Of particular interest are ligonucleotidetemplated reactions that can be monitored with high sensitivity by the appearance/disappearance of a fluorescent signal upon binding to the oligonucleotide target [15-17]. Representative examples of such technologies include the use of fluorogenic probes (e.g. molecular beacons [18,19], or rely on fluorogenic reactions of chemical ligation [20] or primer extension [21]. Most recent reports are based on the Staudinger reaction [22-27] diamine-catalysed aldol condensation [28] organomercury-activated [29] or SNAr reactions [30]. Among these examples, a small proportion only was sucessfully applied for the detection of SNPs in vitro. Briefly, two modified oligonucleotides (or oligonucleotide analogues) are designed so that they can hybridize in a sequence-specific manner to a unique nucleic acid template through Watson-Crick base pairing.Upon hybridization of the synthetic probes to the complementary DNA strand, both fluorogenic moieties are brought in close enough proximity to react with each other, thus generating the fluorescent product. In case of a partial or uncomplete hybridization of one of the modified oligonucleotides (e.g. as a consequence of a SNP), both probes are no longer positioned favorably to react with each other, thus leading to a significant decrease of reaction efficiency (i.e. a weaker fluorescence). Within such systems, genetic information can be directly linked to the appearance of a characteristic fluorescence signal.Herein, we report a biocompatible and fluorogenic system that uses two water-soluble PNAs functionalised at their N- or C- terminus with non-fluorescent cyanine dye precursors for SNPs detection. Briefly, two 5-mer PNAs were synthesized on solid support that also contained two ε-N,N-dimethyl-Lysines to ensure water-solubility and were functionalised either at their N- or C-terminus with a Fischer’s base aldehyde and 2-methylene-indolenine, respectively [31,32]. Upon simultaneous hybridization of both PNAs to a complementary DNA template only, both fluorogenic probes will be orientated optimally to react with each other, thus producing irreversibly a highly fluorescent symmetrical trimethine cyanine dye (Figure 1).
Figure 1: General strategy for SNPs detection using two PNAs functionalised with non-fluorescent trimethine cyanine dye precursors.
Materials and Methods
General
Sensing experiments
Stock solutions (50 μM) of Pna1, Pna2 and DNA were prepared in water. In a typical experiment, 20 μL of a potassium phosphate buffer solution (100 mM, pH = 7.4) and 10 μL of each stock solution were transferred into one well to make the final concentrations of 10 μM Pna1, 10 μM Pna2, 10 μM DNA and 40 mM buffer. Reaction time-courses were determined in a 384-well plate using a SpectraMax M5 fluorescence plate reader (Molecular devices, UK). Reactions were carried out at 37°C with λexc = 540 nm and λem = 573 nm.Results and Discussion
The ability of Pna1 and Pna2 to react with each other in the absence and in the presence of various DNA sequences used as templates was investigated by fluorescence spectroscopy by monitoring the formation of a symmetrical trimethine cyanine dye (λexc = 540 nm, λem = 573 nm). Reactions were carried out in potassium phosphate buffer (40 mM, pH 7.4) at 37°C using a stoichiometric mixture of Pna1, Pna2 and DNA (10 μM each, total volume 50 μL) and the efficiency of the fluorogenic reaction was monitored over three hours in a fluorescence microplate reader. Importantly, no fluorescence was ever detected when incubating Pna1 and Pna2 under such conditions of concentration and pH and in the absence of any DNA template (Figure 3, grey curve).Figure 3: Reaction time-course of a stoichiometric mixture of Pna1 and Pna2 (10 μM each) in the absence (grey) and in the presence of a stoichiometric amount of WT0 (blue), WT1 (red), WT2 (black), WT3 (green) or WT4 (pink). Reaction was monitored by fluorescence spectroscopy (λexc = 540 nm, λem = 573 nm).
Table 1: Oligonucleotide sequences (given from the 5’ to 3’ end)a.
Name | Sequence | Name | Sequence |
WT0 | GCATCTCGGC | Mut5 | GCATCCTTCGGT |
WT1 | GCATCTTCGGC | Mut6 | GCATCCTTCGGC |
WT2 | GCATCCTTCGGC | Mut7 | GCATCCTTXCGGCb |
WT3 | GCATCCTTTCGGC | Mut8 | GCATXCTTTCGGCb |
WT4 | GCATCCTTTTCGGC | Mut9 | GCATCCTTAAGGC |
Mut1 | GCATCCTXCGGCb | Mut10 | GCATCCTTAAAGC |
Mut2 | GCATCCTTTGGC | Mut11 | GCATACTTACGGC |
Mut3 | GCATCCTTCTGC | Mut12 | GCAAACTTAAGGC |
Mut4 | GCATCCTTCGTC | ||
| aUnderlined sequences are that complementary to Pna1 and Pna2;bXrepresents A, T, C or G. | |||
Figure 4: Reaction time-course of a stoichiometric mixture of Pna1 and Pna2 (10 μM each) in the presence of a stoichiometric amount of WT3 (black) or one of the mutated sequences Mut7G (red), Mut7C (blue), Mut7A (green), Mut8G (pink) or Mut8A (grey) (Fig. 4A), or Mut9 (red), Mut10 (blue), Mut11 (green) or Mut12 (grey) (Fig. 4B). Reaction was monitored by fluorescence spectroscopy (λexc = 540 nm, λem = 573 nm).
Therefore, and despite the fact that WT2, as a template, was not quite as efficient as WT3, we reasoned that a 2-nucleotide gap could provide a more rigid environment for the fluorogenic reaction so that the correct alignement of both non-fluorescent cyanine dye precursors would be more responsive to SNPs. DNA sequences derived from WT2 but carrying single nucleotide mutations at various positions (Mut1-6, Table 1) were tested for their ability to template the fluorogenic reaction. The reaction time-courses were determined by fluorescence spectroscopy on a microtiter plate format and compared to that obtained with the corresponding nativesequence WT2 (Figure 5).
Figure 5: Reaction time-course of a stoichiometric mixture of Pna1 and Pna2(10 μM each) in the presence of a stoichiometric amount of WT2 (black), Mut1 (red), Mut2 (blue), Mut3 (grey), Mut4 (pink), Mut5 (green). Reaction was monitored by fluorescence spectroscopy (λexc = 540 nm, λem = 573 nm).
Finally, in order to estimate the efficiency of the fluorogenic reaction upon addition of complementary DNA, a water-soluble analogue of the symmetrical trimethine cyanine dye reaction product was synthesized and used as a standard for quantifying the amount of dye formed in the absence or presence of various DNA templates. Using this external standard, the yield of the reaction templated by WT2 was found to be approximately 10 ± 5 % while no fluorescent product was ever detectable under similar conditions but in the absence of template (Figure 6). Interestingly, addition of a large excess (20 μg/mL) of competitor Calf-Thymus DNA in the reaction mixture resulted in no significant loss of reaction efficiency (data not shown), thus confirming the high specificity of our fluorogenic system.
Figure 6: Fluorescence calibration curve (fluorescence intensity 25 versus dye concentration, λexc = 540 nm, λem = 573 nm) obtained from an analytically pure sample of symmetrical trimethine cyanine dye similar to the product of the fluorogenic reaction (inset). The fluorescence intensity obtained at equilibrium (after 3 hours) for a stoichiometric 30 mixture of Pna1 and Pna2 (10 μM each) in the presence of a stoichiometric amount of WT2 is highlighted in red.
Conclusion
In conclusion, we have reported the use of a fast, versatile, biocompatible and fluorogenic DNA-templated reaction of cyanine dye formation for the detection of SNPs in vitro. The system offers the advantage of a reasonably high sensitivity due to the high absorptivity and moderately high fluorescence quantum yield of the cyanine dye formed [33] and requires minimal amounts (500 pmol) of both DNA and PNA probes when the reaction is carried out in a 384-well microtiter plate. The design of optimized (e.g. more rigid, multicoloured) systems to further improve the sensitivity of the detection is currently underway in our laboratory.References
- Collins FS, Green ED, Guttmacher AE, Guyer MS (2003) A vision for the future of genomics research. Nature 422: 835-847.
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. (2001) The sequence of the human genome. Science 291: 1304-1351.
- Stankiewicz P, Lupski JR (2010) Structural variation in the human genome and its role in disease. Annu Rev Med 61: 437-455.
- Feuk L, CarsonAR, Scherer SW (2006) Structural variation in the human genome. Nat Rev Genet 7: 85-97.
- Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, et al. (2000) An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407: 513-516.
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, et al. (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet 22: 239-247.
- Imyanitov EN (2009) Gene polymorphisms, apoptotic capacity and cancer risk. Human Genet 125: 239-246.
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al.(2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851-861.
- Kruglyak L, Nickerson DA (2001)Variation in the spice of life. Nat Genet 27: 234-236.
- Ding C, Jin S (2009) High-throughput methods for SNP genotyping. Methods Mol Biol 578: 245-254.
- LaFramboise T (2009) Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res 37: 4181-4193.
- Kim S, Misra A (2007) SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng 9: 289-320.
- Percivalle C, Bartolo JF, Ladame S (2013) Oligonucleotide-templated chemical reactions: pushing the boundaries of a nature-inspired process. Org Biomol Chem 11: 16-26.
- Gorska K, Winssinger N (2013) Reactions templated by nucleic acids: more ways to translate oligonucleotide-based instructions into emerging function. Angew Chem Int Ed Engl 52: 6820-6843.
- Shibata A, Uzawa T, Nakashima Y, Ito M, Nakano Y, et al. (2013) Very rapid DNA-templated reaction for efficient signal amplification and its steady-state kinetic analysis of the turnover cycle. J Am Chem Soc 135: 14172-14178.
- Sadhu KK, Winssinger N (2013) Detection of miRNA in live cells by using templated RuII-catalyzed unmasking of a fluorophore. Chemistry 19: 8182-8189.
- Ranasinghe RT, Brown T (2005) Fluorescence based strategies for genetic analysis. Chem Commun 5487-5502.
- Venkatesan N, SeoYJ, Kim BH (2008) Quencher-free molecular beacons: a new strategy in fluorescence based nucleic acid analysis. Chem Soc Rev 37: 648-663.
- Marras SA, Kramer, FR, Tyagi S (2003) Genotyping SNPs with molecular beacons. Methods Mol Biol 212: 111-128.
- Toubanaki DK, Christopoulos TK, Ioannou PC, Flordellis CS (2009) Identification of single-nucleotide polymorphisms by the oligonucleotide ligation reaction: a DNA biosensor for simultaneous visual detection of both alleles. Anal Chem 81: 218-224.
- Krieg A, Laib S, Ruckstuhl T, Seeger S (2004) Fast detection of single nucleotide polymorphisms (SNPs) by primer elongation with monitoring of supercritical-angle fluorescence. Chem Bio Chem 5: 1680-1685.
- Cai J, Li X, Yue X, Taylor JS (2004) Nucleic acid-triggered fluorescent probe activation by the Staudinger reaction. J Am Chem Soc 126: 16324-16325.
- Pianowski ZL, Winssinger N (2007) Fluorescence-based detection of single nucleotide permutation in DNA via catalytically templated reaction. Chem Commun 3820-3822.
- Franzini RM, Kool ET (2008) 7-Azidomethoxy-coumarins as profluorophores for templated nucleic acid detection. Chembiochem 9: 2981-2988.
- Pianowski Z, Gorska K, Oswald L, Merten CA, Winssinger N (2009) Imaging of mRNA in live cells using nucleic acid-templated reduction of azidorhodamine probes. J Am Chem Soc131: 6492-6497.
- Furukawa K, Abe H, Hibino K, Sako Y, Tsuneda S, et al. (2009) Reductiontriggered fluorescent amplification probe for the detection of endogenous RNAs in living human cells. Bioconjug Chem 20:1026-1036.
- Saneyoshi S, Ito Y, Abe H (2013) Long-lived luminogenic probe for detection of RNA in a crude solution of living bacterial cells. J Am Chem Soc 135: 13632-13635.
- Huang Y, Coull JM (2008) Diamine catalyzed hemicyanine dye formation from nonfluorescent precursors through DNA programmed chemistry. J Am Chem Soc130: 3238-3239.
- Franzini RM, Kool ET (2008) Organometallic activation of a fluorogen for templated nucleic acid detection.Org Lett 10: 2935-2938.
- Shibata A, Abe H, Ito M, Kondo Y, Shimizu S, et al. (2009) DNA templated nucleophilic aromatic substitution reactions for fluorogenic sensing of oligonucleotides. Chem Commun 6586-6588.
- Meguellati K, Koripelly G, Ladame S (2010) DNA-templated synthesis of trimethine cyanine dyes: a versatile fluorogenic reaction for sensing G-quadruplex formation. Angew Chem Int Ed 49: 2738-2742.
- Koripelly G, Meguellati K, Ladame S (2010) Dual sensing of hairpin and quadruplex DNA structures using multicolored peptide nucleic acid fluorescent probes. Bioconjugchem 21: 2103-2109.
- Benson RC, Kues HA (1977) Absorption and fluorescence properties of cyanine dyes. J Chem Eng Data 22: 379-383.