Journal of Veterinary Science & Medicine
Download PDF
Research Article
*Address for Correspondence: Huaguang Lu, Animal Diagnostic Laboratory, Department of Veterinary and Biomedical Sciences, The Pennsylvania State University, University Park, PA 16802, USA, Tel: 814-863-4369; Fax: 814-865-4717; E-mail: hxl15@psu.edu
Citation: Tang Y, Lin L, Knoll EA, Dunn PA, Wallner-Pendleton EA, et al. The σC Gene Characterization of Seven Turkey Arthritis Reovirus Field Isolates in Pennsylvania during 2011-2014. J Veter Sci Med. 2015;3(1): 7.
Copyright © 2015 Lu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Veterinary Science & Medicine | ISSN: 2325-4645 | Volume: 3, Issue: 1
Submission: 17 March 2015 | Accepted: 22 April 2015 | Published: 27 April 2015
Reviewed & Approved by: Dr. Leyi Wang, Animal Diagnostic Laboratory, Ohio Department of Agriculture, USA
TARV field isolates
RNA extraction and RT-PCR
Sequence of σC protein alignment
Comparison analyses of σC gene
Division of σC sequences
The σC Gene Characterization of Seven Turkey Arthritis Reovirus Field Isolates in Pennsylvania during 2011-2014
Yi Tang, Lin Lin, Eric A. Knoll, Patricia A. Dunn, Eva A. Wallner-Pendleton and Huaguang Lu*
- Animal Diagnostic Laboratory, Department of Veterinary and Biomedical Sciences, The Pennsylvania State University, University Park, PA 16802, USA
*Address for Correspondence: Huaguang Lu, Animal Diagnostic Laboratory, Department of Veterinary and Biomedical Sciences, The Pennsylvania State University, University Park, PA 16802, USA, Tel: 814-863-4369; Fax: 814-865-4717; E-mail: hxl15@psu.edu
Citation: Tang Y, Lin L, Knoll EA, Dunn PA, Wallner-Pendleton EA, et al. The σC Gene Characterization of Seven Turkey Arthritis Reovirus Field Isolates in Pennsylvania during 2011-2014. J Veter Sci Med. 2015;3(1): 7.
Copyright © 2015 Lu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Veterinary Science & Medicine | ISSN: 2325-4645 | Volume: 3, Issue: 1
Submission: 17 March 2015 | Accepted: 22 April 2015 | Published: 27 April 2015
Reviewed & Approved by: Dr. Leyi Wang, Animal Diagnostic Laboratory, Ohio Department of Agriculture, USA
Abstract
Seven turkey arthritis reovirus (TARV) field isolates made in our laboratory during 2011-2014 were characterized by sequencing of reovirus σC genes. Of the seven TARV field isolates, six were isolated from 3-to-17-week-old turkeys with tenosynovitis in six Pennsylvania farms and one was isolated from an Indiana turkey case submitted to our lab in 2014. This report describes the amplification and sequencing of the σC genes for genetic characterization studies on the seven TARV field isolates. Phylogenetic analysis of the sequence data of the seven TARV isolates together with recent published five TARVs detected in Minnesota (MN) and 25 avian reovirus (ARV) strains retrieved from GenBank revealed that all the seven PA TARV field isolates and five MN TARVs fit into genotyping cluster two when compared with a total of five different genotyping clusters (cluster 1-5) generated by the TARV and ARV reference strains. Comparison of amino acidsequences of the seven TARV isolates in cluster two with ARV vaccine strains (S1133, 1733, and 2048) in cluster one revealed that there were less than 60% similarity in nucleotide sequence and less than 56% in amino acid sequence between the two clusters. However the seven PA TARV isolates shared greater than 99% similarity with each other. Our research findings have indicated that the seven PA TARV field isolates and the five MN TARVs are grouped in the same genotype two, a separate genotype or virus species within the Orthoreovirus genus.Keywords
Turkey arthritis reovirus; Reovirus σC genes; Genotype; PhylogeneticAbbreviations
aa: Amino acids; nt: Nucleotide; ARV: Avian Reoviruses; bp: Base Pair; CPE: Cytopathic Effects; LMH: Male-Chicken Hepatocellular- Carcinoma; ORF: Open Reading Frame; RT: Reverse Transcriptase; TARV: Turkey Arthritis ReovirusIntroduction
Avian reoviruses (ARV) belong to the genus Orthoreovirus in the family Reoviridae. They are non-enveloped viruses and contain a double-stranded RNA genome with ten segments. The viral genome is enclosed within a double protein capsid shell with a diameter of 70-80 nm [1,2]. Based on migration pattern on polyacrylamide gel electrophoresis, the ten genomic segments can be separated into three large segments (L1, L2, L3), three medium segments (M1, M2, M3), and four small segments (S1, S2, S3, S4) [3,4]. The segmented genome encodes for at least eight structural proteins (λA, λB, λC, μA, μB, σA, σB and σC) and four non-structural proteins (μNS, P10, P17 and σNS) [5].The ARV σC protein is a minor outer-capsid protein and is encoded by the largest open reading frame (ORF) of the S1 genomic segment. Although the S1 genomic segment is relatively small protein of 327 aa, it plays an important role for virus attachment [6] and acts as apoptosis inducer [7]. Thenucleotide (nt) homology and aa homology of σC have been found suitable for comparison among different strains; firstly, the σC protein is the most variable protein in the ARV [8,9] for both very hypervariable aa regions one to 122 and 196 to 326 [10]; and secondly, it induces the production of neutralizing antibodies [11].
ARV strains have been associated with disease conditions such as viral arthritis/tenosynovitis [12] and can also cause damage to liver, heart and intestine [13]. All avian species of domestic poultry and wild birds are very susceptible to ARV infections, especially when they are young [14]. ARV infections have been reported in various avian species including chickens [15], geese [16,17], turkeys [18,19], ducks [20-22], pigeons [23], quail [24-26], and psittacine birds [27]. However, meat-type chickens have been shown to bemore susceptible to ARV infection than other avian species (De Gussem et al., 2010; Jones and El-Taher, 1985). ARV infections in turkeys are less well understoodwhen compared to ARV infections in chickens.
Turkey arthritis reovirus (TARV) infections were initially reported in 1980’s in the United States [28,29]. Thereafter, no TARV case reports until the recent TARV out breaks occurred in commercial turkeys in the Midwest of the United States in 2009 and 2010 [30]. In Pennsylvania (PA), there are about four commercial companies producing several hundred flocks of turkeys, with average flock size around 12,000 birds per flock. TARV infections in turkeys were diagnosed the first time in PA in June 2011. Two commercial turkey flocks from one company were initially affected with severe lameness and swollen leg joins or entire legs. The causative agent of TARV was isolated from tendon and synovial tissues in the two affected turkey flocks, which represented the first confirmed TARV infections in the East of the United States. Similar TARV infections with various morbidities (30-40%) and mortalities (5-10%) were diagnosed continuously in PA turkey flocks till the present time. A total of 62 TARV field isolates were obtained during 2011 and 2014, and a turkey company had an estimated $3 million dollar of losses in 2014. In the present study, we report our research findings on genetic characterizations of seven TARV field isolates obtained from six PA turkey flocks/farms and one Indiana (IN) turkey flock during 2011-2014, and comparisons of the σC gene nt sequences of the seven TARV field isolates with other reported TARVs and ARV reference strains.
Materials and Methods
A total of seven TARVs isolated from clinical cases in our laboratory during 2011 to 2014 were selected for genetic characterization studies. Of the seven TARV isolates, six were isolated from six different turkey farms in Pennsylvania and one was isolated from samples submitted from an IN turkey flock. All seven turkey flocks had clinical signs of lameness and tenosynovitis (Table 1) and the TARVs were isolated from tendons. Virus isolation and propagation were conducted in Leghorn Male-chicken Hepatocellular-carcinoma (LMH) (CRL- 2113, ATCC, Manassas, VA) cell cultures as per routine cell culture procedures [31]. The TRAV infected LMH cell cultures showed giant or bloom-like cytopathic effects (CPE). When 70-100% CPEs were observed, the LMH cell culture flask was frozen-thawed 2-3 times, and then the cell culture materials were transferred to a 15 ml sterile centrifuge tube for centrifugation at 2000 g for 10 min, and thereafter the supernatant was collected for this research study.
The RNA extraction was carried out with an RNeasy Mini Kit (Cat. No.74106, QIAGEN, Valencia, CA) following the manufacturer’s instructions. Ten microliters of RNA solution were used for synthesis of viral cDNA using the One Step RT-PCR Kit (Cat. No. 210212, QIAGEN, Valencia, CA) with two degenerate primers P1 (5’-AGTATTTGTGAGTACGATTG-3’) and P4 (5’-GGCGCCACACCTTAGGT-3’) which corresponding to σC gene of ARV [32]. The RT-PCR reaction master mix per reaction consists of 25 μl of RNase-free water, 10 μl of 5× Buffer, 2 μl of dNTP mix (10 mM each dNTP), 1 μl Enzyme mix, and 1 μl of each of the two primers (20 pMol/μl), which brings a total volume of 40 μl of the RTPCR master mix. The thermal cycling parameters are set as, one cycle for RT step at 50 °C for 30 min, initial PCR activation step at 95 °C for 15 min, and then followed by 38 cycles for PCR step at 94 °C for 30s for denaturation, 50 °C for 30s for annealing, 72 °C for 90s for extension, 72 °C for 5 min for final extension.
RT-PCR product purification and sequencing
RT-PCR products were isolated by 1% agarose gel electrophoresis and purified by gel extraction kit (Lot No. 04113KE1, Axygen, Tewksbury, MA) following the manufacturer’s instructions. The DNA concentration of the purified PCR product was measured using a NanoDrop™1000 (Thermo Scientific, Waltham, MA) spectrophotometer and then submitted to Penn State Genomics Core Facility for Sanger sequencing.
Phylogenetic analysis
Phylogenetic analysis was performed on nt of the σC gene (981 bases) in S1 segment (525–1613, 1088 bases). Sequence data from all isolates were edited using DNAMAN software (version 7.212, Lynnon Corp., San Ramon, CA). Prediction of the ORFs and translation of ORF into aa sequences were also conducted by using the DNAMAN software. Sequence similarities of the seven TARV field isolates with other avian reovirus reference strains were analyzed by BLAST search in GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and their sequences were aligned by using the ClustalW 1.83 program (http://align.genome.jp/). The aa and nt identities were obtained using ClustalW matrix as the comparison scoring tables for aa and nt comparisons, respectively. Phylogenetic trees were constructed using neighbor-joining method implemented in MEGA4.0 [33]. Bootstrap analysis was performed with 1000 pseudoreplicates.
Results
Prediction and comparison of aa (1 to 286) sequences of σC segments from the seven PA TARV field isolates, five MN TARV strains, and standard ARV vaccine strains (Figure 1) revealed that aa residues are highly conserved between the seven PA TARVs and five MN TARVs (aa identity>98.6%). The TARVs and ARV vaccine trains shared two high genetic variability sites in the N-terminal portion (aa 41 to 120) and the C-terminus portions (aa 233 to 266). The C-terminal portions of σC monomers of the seven PA TARVs and five MN TARVs contained a number of universally conserved aromatic aa residues (aa 267 to 286). The single aa variation among PA TARVs and MN TARVs were found in residues, 13, 24, 26, 30, 33, 38, 77, 105, 137, 212, 238 and 239.
Comparison analyses of σC gene
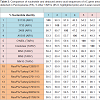
BLAST results showed that all the seven PA TARV shared high sequences similarities with each other (nt: 98.8–99.8%; aa: 97.6-99.7%), but from 47.4% to 99.8% nt similarities with other reference ARV sequences published in GenBank. Pairwise comparison indicated that the nt sequences of the σC protein of the seven PA TARV isolates exhibited high identity with those of the MN TARV field strains (nt: 98.5–99.7%; aa: 98.3–99.7%) that were isolated in MN in 2011 [30]. When compared with ARV S1133, 1733, 2048, and AVS-B strains, the σC sequences of the seven PA TARV isolates shared low similarity to the ARV reference strains (nt, 55.0–59.9%; aa, 48.3–55.2%) (Table 3). Since the TRAVs are in genotyping cluster two, and the three vaccine stains of S1133, 2408 and 138 in genotyping cluster one are very similar (nt>95%) each other (Figure 1), thus we used two (S1133 and 2408) of the three vaccine strains in Table 3 and Figure 2 for aa comparisons.
Figure 2: Phylogenetic tree showing the five clusters of ARV isolates. The analysis was based on the sequence of σC protein. Branch lengths are proportional to the evolutionary distances between sequences. The scale representing nt substitutions perposition is shown. The sequences were either retrieved from GenBank or sequenced in this study. The accession numbers are given in the “Material and methods” section.
Construction of phylogenetic-tree analysis for conservation of the seven TARV σC sequences with other 30 ARV sequences retrieved from GenBank, resulted in five clusters based on their σC sequences (Figure 2), which showed more than 70% identity within each cluster. The ARV vaccine strains (S1133, 1733 and 2048) grouped into cluster onewere very diverse from all of the seven PA TARVs and 5 MN TARVs in cluster two (Figure 2). There are three chicken-origin ARV reference strains (916, ISR528, GEL13A 98M) were felled into the cluster two, only one (GEL13a98M) [32] showed a close relationship to TARVs but with more than 14% nt and 13% aa divergence.
GenBank accession numbers
Sequences of the seven TARV field isolates were submitted to GenBank and published recently, and their assigned GenBank accession numbers are: KM116023 for Reo/PA/Turkey/12883/11, KM116022 for Reo/PA/Turkey/13417/11, KM116019 for Reo/ PA/Turkey/27011/13, KM116020 for Reo/PA/Turkey/22342/13, KM116021 for Reo/PA/Turkey/17010/13, KM116024 for Reo/PA/ Turkey/00659/14, and KM116025 for Reo/Turkey/PA/01769/14 (Table 2).
Table 3: Comparison of nucleotide and deduced amino acid sequences of σC gene among the 7 turkey arthritis reovirus (TARV) isolates (#10-16, highlighted origin) detected in Pennsylvania (PA), 5 other TARVs (#5-9, highlighted pink) and 4 avian reovirus (ARV) (#1-4, highlighted blue) vaccine strains retrieved from GenBank.
Discussion
Research studies have suggested that the σC gene diverges more quickly than the other S-class genes due to the selection pressure placed on a cell-attachment protein [34]. ARV σC protein plays an important role in virus attachment to host cells and as apoptosis inducer. Exchange of σC protein gene segments would enable the virus to continue circulating [9]. In this study, the alignment of σC gene sequences showed that the seven TARV field strains were evolutionarily distant from the three traditional reference ARV strains of S1133, 1733 and 138 as they shared only about 61% nt identity and 54% aa identity. The seven TARV sequences examined in this study had high nt and aa identities not only with each other but also with the five MN turkey reovirus field strains isolated in 2011. This finding suggests that the recent PA TARV may have originatedfrom the Midwest.Among all 12 single aa variation sites of the seven PA and five MN TARVs, only four out of the 12 sites were displayed by two or multiple strains, but the other eight sites were limited to each site by a single strain, which suggests that the aa substitutions may be continuing in the σC gene of the re-emerging TARVs. Continued investigation may reveal more changes in the σC encoding gene that help understand the epidemiology of these infections. Traditionally, ARV strain were classified by virus neutralization tests and the conventional RTPCR in combination with sequencing or other molecular techniques [35,36]. The ARV σC protein is an outer-capsid, mediating virus attachment to cells, and containing antigenic epitopes that can induce neutralizing antibodies [6]. The σC gene displayed the highest level of sequence divergence and rapid evolution; therefore, the gene could be used as a genetic marker for rapid differentiation and classification of TARV field strains or isolates. By working on the full-length σC encoding gene of ARV, which is responsible for serotype specificity in ARV, it is possible to obtain a better correlation between genetic and serologic classification since no correlative relationship has been found between genotypes, serotypes, and pathotypes [32].
Phylogenetic analysis of the seven PA TARV sequences was compared with those of other 30 ARV strains or isolates documented in publications on the basis of the σC gene. The results revealed that the nt of the σC gene had variability, with a maximum divergence of 45% at nt level and 53% at deduced aa level. The σC gene of the seven PA TARV field strains and reference strains had formed five distinct genotyping clusters, while the other S-class genes were reported to have diverged into two or three distinct lineages [9]. The seven PA TARVs in this study and five MN TARVs were all categorized in one group at genotyping cluster two. Despite the close relationship with the five MN TARVs belonging to the same cluster; 916, ISR528, and GEL13A98M strains isolated from chickens with malabsorptionsyndrome were also in the same cluster. The GEL13A98M shared 76% nt similarity with turkey isolates of this study, which is in agreement with previous studies reporting 73.6%–83.1% nt similarity between enteric reoviruses of chickens and turkeys [37]. This finding suggests that enteric reovirus and tenosynovitis reoviruses in turkeys may have originated from a common ancestor. To better understand the relationship between these groups of reoviruses, full genomic sequencing of TARV along with pathogenicity studies should be conducted to obtain more detail information.
Acknowledgements
This research was funded by the Pennsylvania Department of Agriculture, Animal Health and Diagnostic Commission’s project in 2013-2014, and the Pennsylvania Soybean Board’s research project in 2015.References
- Spandidos DA, Graham AF (1976) Physical and chemical characterization of an avian reovirus. J Virol 19: 968-976.
- van der Heide L (2000) The history of avian reovirus. Avian Dis 44: 638-641.
- Nick H, Cursiefen D, Becht H (1975) Structural and growth characteristics of two avian reoviruses. Arch Virol 48: 261-269.
- Varela R, Benavente J (1994) Protein coding assignment of avian reovirus strain S1133. J Virol 68: 6775-6777.
- Varela R, Martinez-Costas J, Mallo M, Benavente J (1996) Intracellular posttranslational modifications of S1133 avian reovirus proteins. J Virol 70:2974-2981.
- Martínez-Costas J, Grande A, Varela R, García-Martínez C, Benavente J (1997) Protein architecture of avian reovirus S1133 and identification of the cell attachment protein. J Virol 71: 59-64.
- Shih WL, Hsu HW, Liao MH, Lee LH, Liu HJ (2004) Avian reovirus sigmaC protein induces apoptosis in cultured cells. Virology 321: 65-74.
- Liu HJ, Giambrone JJ (1997) In situ detection of reovirus in formalin-fixed, paraffin-embedded chicken tissues using a digoxigenin-labeled cDNA probe. Avian Dis 41: 447-451.
- Liu HJ, Lee LH, Hsu HW, Kuo LC, Liao MH (2003) Molecular evolution of avian reovirus: evidence for genetic diversity and reassortment of the S-class genome segments and multiple cocirculating lineages. Virology 314: 336-349.
- Guardado Calvo P, Fox GC, Hermo Parrado XL, Llamas-Saiz AL, Costas C, et al. (2005) Structure of the carboxy-terminal receptor-binding domain of avian reovirus fibre sigmaC. J Mol Biol 354: 137-149.
- Wickramasinghe R, Meanger J, Enriquez CE, Wilcox GE (1993) Avian reovirus proteins associated with neutralization of virus infectivity. Virology 194: 688-696.
- Wilcox GE, Robertson MD, Lines AD (1985) Adaptation and characteristics of replication of a strain of avian reovirus in Vero cells. Avian Pathol 14: 321-328.
- Robertson MD, Wilcox GE, Kibenge FS (1984) Prevalence of reoviruses in commercial chickens. Aust Vet J 61: 319-322.
- Rosenberger JK, Sterner FJ, Botts S, Lee KP, Margolin A (1989) In vitro and in vivo characterization of avian reoviruses. I. Pathogenicity and antigenic relatedness of several avian reovirus isolates. Avian Dis 33: 535-544.
- Howell SH, Walker LL (1972) Synthesis of DNA in toluene-treated Chlamydomonas reinhardi (DNA replication-chloroplast DNA-cell cycle-electron microscopy). Proc Natl Acad Sci U S A 69: 490-494.
- Bezerra DA, da Silva RR, Kaiano JH, Silvestre RV, de Souza Oliveira D, et al. (2012) Detection of avian group D rotavirus using the polymerase chain reaction for the VP6 gene. J Virol Methods 185: 189-192.
- Palya V, et al. (2003) Reovirus identified as cause of disease in young geese. Avian Pathol 32: 129-138.
- Giangaspero M, Vanopdenbosch E, Nishikawa H, Tabbaa D (1997) Prevalence of antibodies against respiratory viruses (parainfluenza virus type 3, respiratory syncytial virus, reovirus and adenovirus) in relation to productivity in Syrian Awassi sheep. Trop Anim Health Prod 29: 83-91.
- Taber R, Alexander V, Whitford W (1976) Persistent reovirus infection of CHO cells resulting in virus resistance. J Virol 17: 513-524.
- Rey ME, D'Andrea E, Calvert-Evers J, Paximadis M, Boccardo G (1999) Evidence for a phytoreovirus associated with tobacco exhibiting leaf curl symptoms in South Africa. Phytopathology 89: 303-307.
- Baroni A, Bertoncin P, D'Aprile PN, Felluga B (1980) Ultrastructural histopathology of chick embryo chorioallantoic membrane infected with an avian reovirus. Avian Pathol 9: 341-354.
- Petek M, D'Aprile PN, Cancellotti F (1973) Biological and physico-chemical properties of the infectious bursal disease virus (IBDV). Avian Pathol 2: 135-152.
- Vindevogel H, Meulemans G, Pastoret PP, Schwers A, Calberg-Bacq CM (1982) Reovirus infection in the pigeon. Ann Rech Vet 13: 149-152.
- Magee DL, Montgomery RD, Maslin WR, Wu CC, Jack SW (1993) Reovirus associated with excessive mortality in young bobwhite quail. Avian Dis 37: 1130-1135.
- Guy JS, Levy MG, Ley DH, Barnes HJ, Gerig TM (1987) Experimental reproduction of enteritis in bobwhite quail (Colinus virginianus) with Cryptosporidium and reovirus. Avian Dis 31: 713-722.
- Ritter GD, Ley DH, Levy M, Guy J, Barnes HJ (1986) Intestinal cryptosporidiosis and reovirus isolation from bobwhite quail (Colinus virginianus) with enteritis. Avian Dis 30: 603-608.
- Sánchez-Cordón PJ, Hervás J, Chacón de Lara F, Jahn J, Salguero FJ, et al. (2002) Reovirus infection in psittacine birds (Psittacus erithacus): morphologic and immunohistochemical study. Avian Dis 46: 485-492.
- Afaleq AA, Jones RC (1989) Pathogenicity of three turkey and three chicken reoviruses for poults and chicks with particular reference to arthritis/tenosynovitis. Avian Pathol 18: 433-440.
- Levisohn S, Gur-Lavie A, Weisman J (1980) Infectious synovitis in turkeys: Isolation of tenosynovitis virus-like agent. Avian Pathol 9: 1-4.
- Mor SK, Sharafeldin TA, Porter RE, Ziegler A, Patnayak DP, et al. (2013) Isolation and characterization of a turkey arthritis reovirus. Avian Dis 57: 97-103.
- Heggen-Peay CL, Cheema MA, Ali RA, Schat KA, Qureshi MA (2002) Interactions of poult enteritis and mortality syndrome-associated reovirus with various cell types in vitro. Poult Sci 81: 1661-1667.
- Kant A, Balk F, Born L, van Roozelaar D, Heijmans J, et al. (2003) Classification of Dutch and German avian reoviruses by sequencing the sigma C protein. Vet Res 34: 203-212.
- 33. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596-1599.
- Steinhauer DA, Holland JJ (1987) Rapid evolution of RNA viruses. Annu Rev Microbiol 41: 409-33.
- Lee LH, Shien JH, Shieh HK (1998) Detection of avian reovirus RNA and comparison of a portion of genome segment S3 by polymerase chain reaction and restriction enzyme fragment length polymorphism. Res Vet Sci 65: 11-15.
- Kort YH, Bourogâa H, Gribaa L, Scott-Algara D, Ghram A (2013) Molecular characterization of avian reovirus isolates in Tunisia. Virol J 10: 12.
- Pantin-Jackwood MJ, Day JM, Jackwood MW, Spackman E (2008) Enteric viruses detected by molecular methods in commercial chicken and turkey flocks in the United States between 2005 and 2006. Avian Dis 52: 235-244.