Journal of Vaccine & Immunotechnology
Download PDF
Research Article
*Address for Correspondence:Jared D. Evans, Johns Hopkins Applied Physics Laboratory, 11100 Johns Hopkins Rd. Laurel, MD 92307, Tel: 240-228-0191; Fax: 443-778-3500; E-mail: jared.evans@jhuapl.eduTed M. Ross, Vaccine and Gene Therapy Institute of Florida, 9801 SW Discovery Way, Port Saint Lucie, FL34987, Tel: 772-345-5693; Fax: 772-345-0625; E-mail: tross@vgtifl.org
Citation: Ross TM, Tang XC, Lu HR, Olagnier D, Kirchenbaum GA, Evans JD. COBRA-Based Dengue Tetravalent Vaccine Elicits Neutralizing Antibodies Against All Four Dengue Serotypes. J Vaccine Immunotechnology. 2014;1(1): 7.
Copyright © 2014 Ross TM, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Vaccine & Immunotechnology | Volume: 1, Issue: 1
Submission: 29 July 2014 | Accepted: 04 September 2014 | Published: 08 September 2014
Reviewed & Approved by: Dr. Yanjin Zhang, University of Maryland, College Park, MD, USA
COBRA-Based Dengue Tetravalent Vaccine Elicits Neutralizing Antibodies Against All Four Dengue Serotypes
Ted M. Ross1,2,3*, Xian-Chun Tang1, Hai-Rong Lu1, David Olagnier3, Greg A. Kirchenbaum3 and Jared D. Evans1,2,4*
- 1Center for Vaccine Research
- 2DepartmentofMicrobiologyandMolecularGenetics, University of Pittsburgh, Pittsburgh, PA15261, USA;
- 3Vaccine and Gene Therapy Institute of Florida, Port St. Lucie, FL34987
- 4Johns Hopkins Applied Physics Laboratory, Laurel, MD92307
*Address for Correspondence:Jared D. Evans, Johns Hopkins Applied Physics Laboratory, 11100 Johns Hopkins Rd. Laurel, MD 92307, Tel: 240-228-0191; Fax: 443-778-3500; E-mail: jared.evans@jhuapl.eduTed M. Ross, Vaccine and Gene Therapy Institute of Florida, 9801 SW Discovery Way, Port Saint Lucie, FL34987, Tel: 772-345-5693; Fax: 772-345-0625; E-mail: tross@vgtifl.org
Citation: Ross TM, Tang XC, Lu HR, Olagnier D, Kirchenbaum GA, Evans JD. COBRA-Based Dengue Tetravalent Vaccine Elicits Neutralizing Antibodies Against All Four Dengue Serotypes. J Vaccine Immunotechnology. 2014;1(1): 7.
Copyright © 2014 Ross TM, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Vaccine & Immunotechnology | Volume: 1, Issue: 1
Submission: 29 July 2014 | Accepted: 04 September 2014 | Published: 08 September 2014
Reviewed & Approved by: Dr. Yanjin Zhang, University of Maryland, College Park, MD, USA
Abstract
Dengue virus (DENV) is the most common arthropod-borne infection in the world. The co-circulation of four serotypes, complex pathogenesis, and potential for antibody-enhanced disease has made vaccine development efforts difficult. To ensure protection and minimize vaccine-related disease augmentation, a DENV vaccine must provide equivalent immunity to all four serotypes. Four different vaccine formulations were evaluated for efficacy and utility. DNA plasmid based and purified subviral particles (SVP) vaccines were designed using prototype sequences as well as consensus algorithms known as computationally-optimized broadly reactive antigen (COBRA). These vaccines were formulated against each individual serotype, as well as tetravalent mixtures and used to inoculate mice. All monovalent vaccines elicited neutralizing antibodies against each of their specific homologous virus. In contrast, only purified versions of tetravalent subviral particle (SVP) elicited high levels of neutralizing antibodies against all four serotypes. All Dengue COBRA VLP vaccines elicit a broadly reactive immune response against all four subtypes of dengue virus. A non-infectious SVP vaccine that induces immune protection against the four DENV serotypes could provide a safer alternative candidate to live attenuated viruses.Keywords
Dengue; VLP; Vaccine; COBRA; Neutralizating antibodyIntroduction
Dengue viruses (DENV) are the most prevalent mosquitoborne infection in the world, causing a range of physical outcomes from asymptomatic to benign febrile illness to life-threatening hemorrhagic disease [1-3]. Currently, there are no commercially available therapeutic interventions to prevent or treat viral infection. DENV are a group of 4 closely related, yet genetically distinct serotypes belonging to the Flaviviridae family [4]. Dengue is a complex disease for which the exact mechanisms of pathogenesis are not fully understood. However, the predominant theory suggests that the majority of primary infections result in neutralizing antibodies and life-long protection against that particular serotype.Secondary infection with a heterotypic DENV serotype increases the likelihood for severe dengue disease, including lifethreatening hemorrhagic disorders and systematic stress [5,6]. Nonneutralizing antibodies may bind the heterotypic DENV serotypes in a second infection and enhance their entry into Fc-receptor bearing cells, thus increasing infection and disease. This antibodydependent enhancement (ADE) is the major challenge facing vaccine development. For this reason, a vaccine against DENV must produce equivalent immunity to all 4 serotypes or risk the potential for severe dengue disease manifestation.
Results from genetically engineered mice indicate that both the innate (e.g., interferon) and the adaptive (B and T cells) immune responses control DENV infection [7]. The production of antibodies provides protection against DENV infection [8] and passive antibody transfer of anti-DENV neutralizing antibodies can prevent or treat lethal infection [9]. The primary target of the neutralizing antibody response is the E protein, which is the most accessible structural glycoprotein on the surface of the virion [10]. Structural analysis of the soluble ectodomain of flavivirus E proteins reveals three domains [11,12]. Even though neutralizing antibodies are generated against epitopes in all three domains, many highly neutralizing antibodies cluster to epitopes in Domain III (DIII) [13].
Several experimental vaccines for humans based upon liveattenuated virus, chimeric virus, purified protein, viral vectors, or DNA plasmids are under development (see reviews [6,14,15]). However, a highly attractive potential vaccine candidate would be a non-infectious virus-like particle, which has been successfully used as a vaccine delivery platform for many viruses, as reviewed in [16].The sub viral particles (SVP) entities present structures physically similar to virions, but lack infectious RNA genome.
Previously, the use of consensus-based immunogen design has been used to address the diversity of isolates in many viral families, including dengue [13,17-25]. Our research group has recently expanded upon the consensus strategy and developed computationallyoptimized broadly reactive antigen (COBRA) strategies to overcome pathogen sequence diversity. Traditional consensus sequences are generated by aligning a population of sequences, then constructing genes consisting of the most common residue at each position. These sequences are expected to effectively capture conserved epitopes and elicit cross-reactive immune responses. The COBRA methodology of antigen design uses multiple rounds of consensus generation to address antigenic diversity in envelope glycoprotein sequences. The COBRA algorithm compiles three consecutive consensus generations to identify the most appropriate vaccine antigen [20]. Candidate influenza vaccines have been previously tested in mice and ferret models [18-20]. In this report, COBRA dengue E proteins were displayed on the surface of a subvirion particle (SVP) and used to vaccinate mice to elicit broadly reactive neutralizing antibodies.
Materials and Methods
Antigen construction and synthesisDengue E nucleotide sequences isolated from human infections were downloaded from the GenBank Database (accession numbers available up on request) [22,26] . Nucleotide sequences were translated into protein sequences using the standard genetic code. Full-length dengue E sequences (100 from each serotype) from 1941 to 2006 were acquired and used for subsequent consensus generations. For each round of consensus generation, multiple alignment analysis was applied and the consensus sequence was generated using Align X (Vector NTI). The final amino acid sequence, termed computationally optimized broadly reactive antigen (COBRA), as well as the prototype E sequences were reverse translated and optimized for expression in mammalian cells, including codon usage and RNA optimization (Gene Art; Regensburg, Germany).This construct was then synthesized and inserted into the pTR600 expression vector. Prototype virus E sequences were used for prototype sequences and included Hawaii (DENV-1), NCG (DENV-2), H87 (DENV-3), and H241 (DENV-4); (accession numbers X76219, M29095, M93130, S66064). Prototype and COBRA DENV E sequences representing all four subtypes of dengue were constructed.
Construction and expression of DNA vaccine plasmids
Constructs labeled SVP-DNA expressed the full-length prM/E reading frame and encoded the prM and E gene segments were cloned in frame with the tPA leader sequence in pTR 600, as previously described [27] (Figure 1). Human embryonic kidney (HEK) 293 Tcells were transfected with 3μg of DNA using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA) and as previously described [27]. Cell culture supernatants were collected 48 hours post-transfection. Approximately 1.5% of sample volume was loaded onto a 10% polyacrylamide/SDS gel and detected as previously described [27].
Concentration of Subviral Particles (SVP)
Following transfection with SVP-DNA, 293T cells were incubated for 72h at 37°C. Supernatants were collected and cell debris was removed by low speed centrifugation followed by vacuum filtration through a 0.22mM sterile filter. SVP preparations were collected via ultracentrifugation (100,000Xg through 20% glycerol, weight per volume) for 4h at 4°C.The pellets were subsequently resuspended in phosphate buffered saline (PBS) and stored at -80°C.
Protein concentration was determined by Micro BCATM Protein Assay Reagent Kit (Pierce Biotechnology, Rockford, IL).
Virus titrations
Prototype virus isolates were used for neutralization assay and included Hawaii (DENV-1), NCG (DENV-2), H87 (DENV-3), and H241 (DENV-4).Viruses were propagated in C6/36 mosquito cells, aliquotted, and stored at -80°C and they were tittered before use as previously described [28,29]. Briefly, Vero cells were infected with 10-fold serial dilutions of virus in 24- well plates. At day 5, the cells were fixed with Methanol/Acetone (Fisher). Focus forming units were detected with MsX Dengue Complex antibody (Millipore, Billerica, MA) and Goat anti-Mouse-HRP secondary antibody (Millipore, Billerica, MA), using TrueBlue TM peroxidase substrate (KPL, Gaithersburg, MD). Infectious foci were counted and recorded as focus-forming units per milliliter of cell media supernatant (FFU/mL).
Vaccination
Female BALB/c mice (n=8 mice per group; aged 6–8 weeks) were purchased from Harlan Sprague Dawley, (Indianapolis, IN). For DNA immunizations, mice were vaccinated with each DNA vaccine plasmid by gene gun (particle bombardment with 2μg DNA coated on gold bullets) and then boosted with the same DNA on weeks 3 and 6. For purified SVP vaccinations, mice were vaccinated via intra peritoneal route with 200μg of SVP protein per vaccination at week 0, 3 and 6. Blood was collected on weeks 5 and 8 post-vaccination, then centrifuged at 6000 rpm for 10 min to separate the serum. Sera were frozen at -80°C. Animals were treated according to the guide lines of the IACUC of the University of Pittsburgh. All the protocols used were approved by the IACUC of the University of Pittsburgh (#1002617).
Enzyme-Linked Immuno Absorbant Assay (ELISA)
Quantitative ELISA detected anti-E specific IgG in serum of SVPs or SVP-DNA vaccinated mice using purified Dengue E proteins (1mg) (Virostat, Portland, ME, USA) as previously described [27]. The O.D. value of the age-matched naïve sera was subtracted from the O.D. values of the anti sera from the vaccinated mice. Results were recorded as the geometric mean titer (GMT) ± the standard error of the mean (S.E.M.).
Plaque Reduction Neutralization Assay (PRNT)
PRNT assays were performed as previously described [30]. Briefly, pooled sera from groups of mice were heat-inactivated at 56°C for 30 minutes. Serum sample dilutions (1:20-1:640) were tested against prototype viruses (DENV-1-4) at a fixed concentration (200 FFU). Virus control wells consisted of similar dilutions of mockvaccinated mouse serum. Serum samples mixed with virus were incubated for one hour at 37°C. The serum-virus mixture was added to Vero cells in duplicate. Virus controls were also serially diluted 2 to 16 fold for PRNT calculation prior to addition to the cells. After 1.5 hours of infection, the serum-virus mixture was removed and cells overlayed with 0.8% methyl cellulose in Opti-Mem media with 2% heat- inactivated FBS. Infections were incubated for 5 days at 37°C and 5% CO2. Cells were fixed with ice-cold methanol:acetone (1:1). The methanol:acetone solution was removed and cells air dried. Cells were rehydrated with PBS. Dengue-specific antibodies (Millipore, USA) were diluted in PBS supplemented with 3% heatinactivated FBS, added to cells, and incubated with rocking. Plates were washed with PBS, followed by incubation with HRP-conjugated goat anti-mouse secondary antibody diluted in PBS supplemented with 3% heat-inactivated FBS. Infectious foci were detected by the addition of TruBlue HRP substrate (KPL, Gaithersburg) according to manufacturer’s instructions. Infectious foci were counted and recorded. The PRNT titers were scored by calculating the highest dilution of sample that inhibited 50% of the foci (PRNT50) in comparison to virus dilution wells without antibody.
 Figure 1: Schematic diagram of constructs and expression of vaccine plasmids. A. A diagram of the DENV construct expressing prM/E segment of the genome is shown (top). B. Supernatants and cell lysates from 293T cells transiently transfected with plasmid DNA were assessed by SDS-PAGE and Western blot. The membranes were probed with the DENV E-specific monoclonal antibody, 9.F.10 or Actin monoclonal antibody as the loading control for cell lysates.
Figure 1: Schematic diagram of constructs and expression of vaccine plasmids. A. A diagram of the DENV construct expressing prM/E segment of the genome is shown (top). B. Supernatants and cell lysates from 293T cells transiently transfected with plasmid DNA were assessed by SDS-PAGE and Western blot. The membranes were probed with the DENV E-specific monoclonal antibody, 9.F.10 or Actin monoclonal antibody as the loading control for cell lysates.
In addition to DNA vaccines, the SVP-DNA was used to transfect mammalian cells for SVP production and purification (Figure 1B). These genomeless SVPs are generated by transiently transfecting pSVP into 293T cells that results in expression of both prM and E proteins. Previous data suggest prM and E proteins self-assemble and secrete from mammalian cells [31,32]. Secreted SVPs were collected by centrifugation and total protein content was determined. Both cell lysates and supernatants were analyzed for the presence of E protein. The data show that each DENV subtype E protein was expressed to the similar levels in cell lysates (Figure 1B, middle panel), but have different levels of secreted SVPs (Figure 1B, top panel). Each purified SVP represented one subtype of DENV and incorporated E proteins.
Antibody IgG Isotypes
DENV Neutralizing Antibody Titers
Results
Construction of DENV Vaccine PlasmidsDNA plasmids were constructed to express subviral particles (SVP) from prM/E cassettes representing all 4 subtypes of DENV with COBRA or wild-type E genes. All of these gene cassettes were cloned directly downstream of a cytomegalovirus promoter. Each plasmid efficiently expressed the appropriate version of the E protein in transiently transfected 293T cells as determined by Western blot of clarified cell supernatant with specific anti-E MAbs (Figure 1). A 65kD protein representing DENV E was detected in supernatants from cells transiently transfected with DNA expressing the COBRA prM/E gene cassette, which produces subviral particles (SVPs) (see below). As expected, mock-transfected or vector-only transfected cell supernatants showed no reactivity with DENV anti-E MAbs 9.F.10 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
 Figure 1: Schematic diagram of constructs and expression of vaccine plasmids. A. A diagram of the DENV construct expressing prM/E segment of the genome is shown (top). B. Supernatants and cell lysates from 293T cells transiently transfected with plasmid DNA were assessed by SDS-PAGE and Western blot. The membranes were probed with the DENV E-specific monoclonal antibody, 9.F.10 or Actin monoclonal antibody as the loading control for cell lysates.
Figure 1: Schematic diagram of constructs and expression of vaccine plasmids. A. A diagram of the DENV construct expressing prM/E segment of the genome is shown (top). B. Supernatants and cell lysates from 293T cells transiently transfected with plasmid DNA were assessed by SDS-PAGE and Western blot. The membranes were probed with the DENV E-specific monoclonal antibody, 9.F.10 or Actin monoclonal antibody as the loading control for cell lysates.Anti-DENV Total IgG antibody responses
BALB/c mice were vaccinated with the SVP-DNA vaccine via gene gun (GG) or with purified SVPs at weeks 0, 3 and 6. On week 8, the anti-DENV antibody titers were determined by ELISA from individual clarified serum samples (Figure 2). End-point dilution titers greater than 1x102 were considered significant. There was no difference between replicates in the assay, resulting in minimal or absent error bars. In general, mice vaccinated with SVP-DNA expressing COBRA E proteins (Figure 2B) had a similar antibody binding profile to mice vaccinated with purified COBRA SVP (Figure 2A). In contrast, COBRA SVP vaccinated mice had higher antibody binding titers compared to pSVP-DNA vaccinated mice (Figure 2B). Mice vaccinated with purified DENV-1 or DENV-2 COBRA SVPs elicited high titers to E proteins from all 4 serotypes (>1:10,000). In contrast, mice vaccinated with purified wild-type SVPs (Figure 2C and D) generally had antibodies to their homologous DENV, but not as high as COBRA antigens. DENV-3 and DENV-4 COBRA SVP samples only recognized the homologous DENV E protein. However, mice vaccinated with tetravalent mixture of purified SVP or a mixture of SVP-DNA elicited antibodies that recognized all 4 DENV E proteins. In general, mice vaccinated with the same vaccine with wild-type E proteins had lower IgG titers and a more restricted pattern of cross-reactivity compared to mice vaccinated with COBRA-based vaccines (Figure 2C and D). These vaccines expressing wild-type E proteins recognized primarily the homologous serotype. Overall, these vaccinations resulted in measurable antibodies against E proteins homologous to E protein in the vaccine with variable cross-reactivity against heterologous E proteins.
Figure 2: Vaccine elicited anti-DENV E antibodies. Total IgG titers were measured by ELISA on DENV E proteins coated plates from mice vaccinated with DNA plasmids or purified SVPs on week 8. Sera from each mouse group were tested against E proteins representing each subtype of DENV or WNV. (A) Purified COBRA SVP; (B) COBRA SVP-DNA; (C) purified wild-type SVP; (D) wild-type SVP-DNA.
To characterize further the immune response elicited by these vaccines, the IgG subtypes of the elicited anti-DENV E antibodies were determined (Figure 3). Gene gun pSVP-DNA vaccination elicited primarily a T-helper (Th)-2 (characterized by IgG1 isotype). IgG2a and IgG2b titers were detectable, but low in DNA vaccine groups. In contrast, mice vaccinated with purified SVPs had high IgG1, IgG2a, and IgG2b antibody titers. Interestingly, IgG3 titers were detected in mice vaccinated with purified SVP, but not in any of the DNA vaccine groups (Figure 3). Similar results were obtained using SVP vaccines with wild-type E proteins (data not shown).
Figure 3: The IgG isotype of the anti-DENV E antisera. The specific IgG isotype was assayed from the individual serum collected at week 8 from each individual mouse per vaccine group at a dilution of 1:100 and reported as specific O.D.*p > 0.05 compared to corresponding isotype of SVP group; **p.0.01 > compared to corresponding isotype of pSVP group.
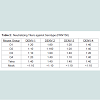
To determine efficacy of the vaccine approaches, neutralizing antibodies against each DENV serotype were measured. Both DNA and protein monovalent SVP vaccines were very effective against homologous virus (Table 1). Further, there was detectable cross reactivity. For example, DENV-2 pSVP-DNA vaccination resulted in neutralizing antibodies to DENV-1, -2, and -4. This pattern was reproduced in the purified DENV-2 SVPs. Vaccination with tetravalent formulations of either plasmid or purified SVP structures yielded similar results. pSVP-DNA and purified SVP vaccinations elicited antibody titers to all 4 serotypes. The standard error for each triplicate of PRNT assay was less than ±10 with many replicates exhibiting identical neutralizing antibody titers (data not shown). However, mice vaccinated with the purified tetravalent-SVP vaccine elicited strongly neutralizing antibodies to each DENV serotype, where as the plasmid-based tetravalent vaccine titers were lower, especially against DENV-3 (Table 1). The neutralizing titers were independent of total antibody levels (Figure 2). Sera collected from animals vaccinated with SVPs containing E protein representing wild-type sequences elicited a lower titer antibody that was more restricted at recognizing E protein across serotypes (Table 2).
Discussion
Dengue virus infections can be asymptomatic or result in life threatening hemorrhagic syndromes [33,34]. Importantly, second heterotypic infections increase the likelihood of more severe disease (For review, see [35]). Therefore, DENV vaccines must elicit equivalent and effective immunity to all four virus serotypes simultaneously. Incomplete protection could result in more severe disease during subsequent infections. In the current study, two different vaccine approaches were evaluated to determine efficacy. While vaccines against individual serotypes produced neutralizing antibody titers against the homologous virus, there was little cross-reactivity toward other serotypes. To this end, a tetravalent SVP vaccine using COBRA designed E proteins was produced. The COBRA vaccine strategy represents a novel method of sequence construction that features a layered building approach that is intended to capture the most common antigenic characteristics, while avoiding the complication of differential sequence availability that can bias a consensus sequence to the most prevalent antigenic cluster [20]. Avoiding this sampling bias is essential to generating a centralized vaccine that accurately represents the population of input sequences. These tetravalent purified COBRA SVPs elicited strong neutralizing antibody titers against all four DENV serotypes (Table 1). Interestingly, the vaccines expressing wild-type SVP sequences elicited very low neutralizing antibody levels (Table 2). The reason for this effect is unclear.Recent studies have produced DENV VLPs and SVPs for potential use as vaccines [36-38]. For other virus systems, such as HIV or influenza, VLPs expressing the envelope glycoproteins are more effective at eliciting high titer neutralizing antibodies than soluble versions [31,38-45]. Genomeless viral particle vaccines, unlike single proteins, can bind appropriate surface receptors and enter cells in a manner similar to true virus infection [45]. Thereby, presenting viral epitopes to T-cells by professional antigen presenting cells [46]. In addition, antibodies can bind to circulating SVPs and be taken up by phagocytic cells via Fc receptors, thus increasing MHC class II presentation [46]. Neutralizing antibodies are often directed against conformational epitopes that are only present in the native envelope form. In contrast, other epitopes are only exposed during viral entry or release. SVPs have the ability to present viral proteins in their natural conformations to elicit antibodies that recognize viral E proteins on the surface of a virion.
In this study, the purified SVP vaccine elicited a different anti-DENV E IgG isotype pattern than the DNA vaccines (Figure 3). The isotype of the polyclonal antibody in part determines the effector functions of the anti-E antibodies and identifies the T helper cell bias (required for antibody classs witching). The predominant isotype elicited by all DNA vaccination immunizations was IgG1, indicating a Th2 bias. For purified SVP vaccination, IgG1, IgG2a, IgG2b, and IgG3 isotypes were detected, indicating that the SVP vaccine elicit a mixed (Th1 and Th2) T-helper response. This response suggests activation of dendritic cells and macrophages. Moreover, murine IgG3 is homologous to human IgG2, the primary protective antibody isotype. Our results demonstrate that SVP protein complex vaccines produce significant levels of IgG3 antibody (Figure 3).
Antibodies of the IgG2a/c and IgG2b subclass fix complement proteins C1q and C3 and can opsonize and inhibit dengue virus infection [47-49]. However, IgG2a/c binds FcgRI with high avidity, facilitating enhanced uptake of virus-antibody complexes by macrophages via antibody dependent enhancement (ADE). It is crucial to note that total antibody titers did not correlate to neutralization of virus infection, suggesting a preponderance of sub-neutralizing antibodies that may interact with infecting virus. Therefore, ADE may be of concern and should be addressed in future studies (Figure 2 and Figure 3; Table 1).
These SVP vaccines have great potential for a number of reasons. First, the virion structures are easily lyophilized, making transport to remote locales easier [50]. Second, the tetravalent make-up of the vaccine allows optimization of the serotype SVP ratio. This approach will induce equivalent antibody levels and effectiveness to all four DENV serotypes. By altering the amount of certain SVP, it will be possible to increase the antibody response to a particular serotype to obtain equivalent immune responses to each serotype. Further, the secretion of SVP was different between serotypes. It is unclear why DENV-1 SVP were secreted more efficiently than DENV-4. This finding is an area where the system can be improved. Third, recombinant SVPs can be synthesized and produced in short order and purified with simple techniques, thus quickly addressing short ages as well as enabling in-need and poorer countries to produce vaccines for their own populations. Finally, since SVPs do not possess genomes, there is no potential for reversion of live-attenuated viruses to infectious phenotypes.
Several approaches have been applied as dengue vaccines in the past [51-53], but there is currently no licensed tetravalent dengue vaccine. The results presented in this study demonstrate that vaccination with SVPs with COBRA E proteins is potent strategy for dengue vaccination that elicits neutralizing antibodies against all four dengue serotypes. COBRA E antigens may possess a combination of antibody epitopes that are not all present in any given wild-type sequence and therefore elicit a unique antibody profile. Further, the tetravalent mixture of SVP with COBRA E proteins complex vaccines was able to elicit significant neutralizing antibody titers after only 3 vaccinations without an adjuvant. Future studies will elucidate the most effective vaccination regimen and potential adjuvant scheme. Currently, there are several vaccines for dengue virus moving through clinical trials [54-56]. A prime-boost vaccination regimen that includes SVPs plus one of these approaches, i.e. live-attenuated, may provide better protection against dengue virus infection than a single vaccine modality only.
COBRA-based vaccines elicited high antibody titers that recognize wild-type prototype DENV E protein from all four serotypes. A tetravalent DENV virus-like particle vaccine elicited higher neutralizing antibody levels than a DNA-based vaccine. These VLP formulations are attractive candidates for human dengue vaccine.
Acknowledgements
This work was supported by an award from the Department of Defense #W81XWH-BAA06-1 to TMR.JDE was supported by funds provided by the Fine Foundation. This project was also funded, in part, under a grant with the Pennsylvania Department of Health. The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions. We would like to thank Matthew Dunn and LaToya Strong for technical assistance. Dengue viruses were obtained from the Biodefense and Emerging Infections Resource.References
- Halstead SB (2007) Dengue. Lancet 370: 1644-1652.
- Lindenbach B, Rice C (2001) Flaviviridae: the viruses and their replication, 4th Edition. Philadelphia: Lippincott, Williams and Wilkins.
- WHO (2006) Scientific Working group Report on Dengue. World Health Organization.
- Halstead SB (2008) Dengue, volume 5. London: Imperial College Press
- Halstead SB, Nimmannitya S, Cohen SN (1970) Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J Biol Med 42: 311-328.
- Thomas SJ, Endy TP (2011) Critical issues in dengue vaccine development. Curr Opin Infect Dis 24: 442-450.
- Samuel MA, Diamond MS (2006) Pathogenesis of West Nile Virus infection: a balance between virulence, innate and adaptive immunity, and viral evasion. J Virol 80: 9349-9360.
- Diamond MS, Shrestha B, Marri A, Mahan D, Engle M (2003) B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J Virol 77: 2578-2586.
- Diamond MS, Sitati EM, Friend LD, Higgs S, Shrestha B, et al. (2003) A critical role for induced IgM in the protection against West Nile virus infection. J Exp Med 198: 1853-1862.
- Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ (2003) Structure of West Nile virus. Science 302: 248.
- Nybakken GE1, Nelson CA, Chen BR, Diamond MS, Fremont DH (2006) Crystal structure of the West Nile virus envelope glycoprotein. JVirol 80: 11467-11474.
- Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, et al. (2006) Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. JVirol 80: 11000-11008.
- Gao F1, Korber BT, Weaver E, Liao HX, Hahn BH, et al. (2004) Centralized immunogens as a vaccine strategy to overcome HIV-1 diversity. Expert Rev Vaccines 3: S161-S168.
- Coller BA, Barrett AD, Thomas SJ (2011) The development of Dengue vaccines. Introduction. Vaccine 29: 7219-7220.
- Coller BA, Clements DE (2011) Dengue vaccines: progress and challenges. Curr Opin Immunol 23: 391-398.
- Saglanmak N, Andreasen V, Simonsen L, Mølbak K, Miller MA, et al. (2011) Gradual changes in the age distribution of excess deaths in the years following the 1918 influenza pandemic in Copenhagen: using epidemiological evidence to detect antigenic drift. Vaccine 29: B42-B48.
- Gao F, Liao HX, Hahn BH, Letvin NL, Korber BT, et al. (2007) Centralized HIV-1 envelope immunogens and neutralizing antibodies.Curr HIV Res 5: 572-577.
- Giles BM, Bissel SJ, Dealmeida DR, Wiley CA, Ross TM (2012) Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin Vaccine Immunol 19(2): 128-139.
- Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, et al. (2012) A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis 205: 1562-1570.
- Giles BM, Ross TM (2011) A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29: 3043-3054.
- McBurney SP, Landucci G, Forthal DN, Ross TM (2012) Evaluation of heterologous vaginal SHIV SF162p4 infection following vaccination with a polyvalent Clade B virus-like particle vaccine. AIDS Res Hum Retroviruses 28: 1063-1072.
- McBurney SP, Ross TM (2009) Human immunodeficiency virus-like particles with consensus envelopes elicited broader cell-mediated peripheral and mucosal immune responses than polyvalent and monovalent Env vaccines. Vaccine 27: 4337-4349.
- Ramanathan MP, Kuo YC, Selling BH, Li Q, Sardesai NY, et al. (2009) Development of a novel DNA SynCon tetravalent dengue vaccine that elicits immune responses against four serotypes. Vaccine 27: 6444-6453.
- Yan J, Corbitt N, Pankhong P, Shin T, Khan A, et al. (2011) Immunogenicity of a novel engineered HIV-1 clade C synthetic consensus-based envelope DNA vaccine. Vaccine 29: 7173-7181.
- Yan J, Yoon H, Kumar S, Ramanathan MP, Corbitt N, et al. Enhanced cellular immune responses elicited by an engineered HIV-1 subtype B consensus-based envelope DNA vaccine. Mol Ther 15: 411-421.
- McBurney SP, Ross TM (2008) Viral sequence diversity: challenges for AIDS vaccine designs. Expert Rev Vaccines 7: 1405-1417.
- Mamelund SE (2011) Geography may explain adult mortality from the 1918-20 influenza pandemic. Epidemics 3: 46-60.
- Tang XC, Lu HR, Ross TM (2010) Hemagglutinin displayed baculovirus protects against highly pathogenic influenza. Vaccine 28: 6821-6831.
- Tang XC, Lu HR, Ross TM (2011) Baculovirus-produced influenza virus-like particles in mammalian cells protect mice from lethal influenza challenge. Viral Immunol 24: 311-319.
- Vasilakis N, Fokam EB, Hanson CT, Weinberg E, Sall AA, et al. (2008) Genetic and phenotypic characterization of sylvatic dengue virus type 2 strains. Virology 377: 296-307.
- Konishi E, Fujii A (2002) Dengue type 2 virus subviral extracellular particles produced by a stably transfected mammalian cell line and their evaluation for a subunit vaccine. Vaccine 20: 1058-1067.
- Russell PK, Brandt WE, Dalrymple JM (1980) Chemical and antigenic structure of flavi viruses. Academic Press, New York.
- WHO (1997) Dengue Haemorrhagic fever: diagnosis, treatment, prevention and control. 2nd edition. Geneva : World Health Organization.
- WHO (2006) Scientific Working group Report on Dengue. World Health Organization.
- Ross TM (2010) Dengue virus. Clin Lab Med 30: 149-160.
- Sugrue RJ, Cui T, Xu Q, Fu J, Chan YC (1997) The production of recombinant dengue virus E protein using Escherichia coli and Pichia pastoris. J Virol Methods 69: 159-169.
- Sugrue RJ, Fu J, Howe J, Chan YC (1997) Expression of the dengue virus structural proteins in Pichia pastoris leads to the generation of virus-like particles. J Gen Virol 78: 1861-1866.
- Zhang S, Liang M, Gu W, Li C, Miao F, et al. (2011) Vaccination with dengue virus-like particles induces humoral and cellular immune responses in mice. Virol J 8: 333.
- McBurney SP, Young KR, Ross TM (2007) Membrane embedded HIV-1 envelope on the surface of a virus-like particle elicits broader immune responses than soluble envelopes. Virology 358: 334-346.
- Swaminathan S, Batra G, Khanna N (2010) Dengue vaccines: state of the art. Expert Opin Ther Pat 20: 819-835.
- Konishi E, Yamaoka M, Kurane I, Mason PW (2000) A DNA vaccine expressing dengue type 2 virus premembrane and envelope genes induces neutralizing antibody and memory B cells in mice. Vaccine 18: 1133-1139.
- Kuwahara M, Konishi E (2010) Evaluation of extracellular subviral particles of dengue virus type 2 and Japanese encephalitis virus produced by Spodoptera frugiperda cells for use as vaccine and diagnostic antigens. Clin Vaccine Immunol 17: 1560-1566.
- Wang PG, Kudelko M, Lo J, Siu LY, Kwok KT, et al. (2009) Efficient assembly and secretion of recombinant subviral particles of the four dengue serotypes using native prM and E proteins. PLoS One 4: e8325.
- Konishi E, Terazawa A, Imoto J (2003) Simultaneous immunization with DNA and protein vaccines against Japanese encephalitis or dengue synergistically increases their own abilities to induce neutralizing antibody in mice. Vaccine 21: 1826-1832.
- Giles BM, Ross TM (2012) Computationally optimized antigens to overcome influenza viral diversity. Expert Rev Vaccines 11: 267-269.
- Jain S, Kamimoto L, Bramley AM, Schmitz AM, Benoit SR, et al. (2009) Hospitalized patients with 2009 H1N1 influenza in the United States, April-June 2009. N Engl J Med 361: 1935-1944.
- Mehlhop E, Ansarah-Sobrinho C, Johnson S, Engle M, Fremont DH, et al. (2007) Complement protein C1q inhibits antibody-dependent enhancement of flavivirus infection in an IgG subclass-specific manner. Cell Host Microbe 2: 417-426.
- Mehlhop E, Nelson S, Jost CA, Gorlatov S, Johnson S, et al. (2009) Complement protein C1q reduces the stoichiometric threshold for antibody-mediated neutralization of West Nile virus. Cell Host Microbe 6: 381-391.
- Yamanaka A, Kosugi S, Konishi E (2008) Infection-enhancing and -neutralizing activities of mouse monoclonal antibodies against dengue type 2 and 4 viruses are controlled by complement levels. J Virol 82: 927-937.
- Chirkova TV1, Naykhin AN, Petukhova GD, Korenkov DA, Donina SA, (2011) Memory T-cell immune response in healthy young adults vaccinated with live attenuated influenza A (H5N2) vaccine. Clin Vaccine Immunol 18: 1710-1718.
- Edelman R, Wasserman SS, Bodison SA, Putnak RJ, Eckels KH, et al. (2003) Phase I trial of 16 formulations of a tetravalent live-attenuated dengue vaccine. Am J Trop Med Hyg 69: 48-60.
- Kitchener S, Nissen M, Nasveld P, Forrat R, Yoksan S, et al. (2006) Immunogenicity and safety of two live-attenuated tetravalent dengue vaccine formulations in healthy Australian adults. Vaccine 24: 1238-1241.
- Sun W, Edelman R, Kanesa-Thasan N, Eckels KH, Putnak JR, et al. (2003) Vaccination of human volunteers with monovalent and tetravalent live-attenuated dengue vaccine candidates. Am J Trop Med Hyg 69: 24-31.
- Durbin AP1, Whitehead SS (2011) Next-generation dengue vaccines: novel strategies currently under development. Viruses 3: 1800-1814.
- Durbin AP, Kirkpatrick BD, Pierce KK, Elwood D, Larsson CJ, et al. (2013) A single dose of any of four different live attenuated tetravalent dengue vaccines is safe and immunogenic in flavivirus-naive adults: a randomized, double-blind clinical trial. J Infect Dis 207: 957-965.
- Sabchareon A1, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, et al. (2012) Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380: 1559-1567.