Journal of Toxins
Download PDF
*Address for Correspondence: Richard A. Manderville, Departments of Chemistry and Toxicology, University of Guelph, Guelph, Ontario, N1G 2W1, Canada, Tel: (519)-824-4120; x53963; Fax: (519)-766-1499; E-mail: rmanderv@uoguelph.ca
Citation: Olaoye OO, Manderville RA. Aptamer Utility in Sensor Platforms for the Detection of Toxins and Heavy Metals. J Toxins. 2017;4(1): 12
Copyright © 2017 Olaoye OO, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Toxins | ISSN: 2328-1723 | Volume: 4, Issue: 1
Submission: 27 February, 2017 | Accepted: 14 March, 2017 | Published: 30 March, 2017
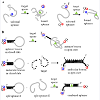
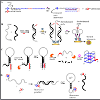
Fluorescence Resonance Energy Transfer (FRET) approach: To effectively overcome the challenges of single fluorophore probes, further studies have designed FRET probes based on fluorescence emission-quenching mechanisms. Bischromophoric approaches tend to show significant increase (>10 folds) in fluorescence intensity on target interactions [46]. FRET aptamer designs attach the fluorophore and quencher at opposite 5′- and 3′- ends of the aptamer sequence, and in a free unbound state, both are separated resulting in maximum fluorescence intensity (Figure 1A, (II)). The aptamer sequence can form a tertiary DNA structure with an introduced target bringing its fluorophore and quencher in close proximity and a fluorescence quenching is observed. This model suffers from the uncertainty in predicting the aptamer structural preference upon target binding. To mitigate this, FRET designs have employed the molecular beacon approach. Molecular beacons can adopt two structures depending on target availability [47,48]. In the absence of desired targets, molecular beacons form an intra molecular stem that places the quencher in close proximity to the fluorophore, thus quenching any emission generated (Figure 1B). When a desired target is however present, molecular beacons can adopt a more stable intermolecular duplex, thus separating the fluorophore-quencher pair and resulting in a fluorescence signal. Sanzani et al. designed an OTA-aptamer loop that readily binds to an OTA target thus releasing and separating the two stems where the fluorophore and quencher are attached [49]. Other variations to the molecular beacon design have also been reported (Figure 1B) [50].
Structure-switching DNAzymes: Based on the reported advantages of aptamers over antibodies [34], several Enzyme-Linked Immunosorbent Assays (ELISA) have incorporated aptamers to act as the target capture probe [65], the detection species [66], or both in some cases in an ELISA variant known as Enzyme-Linked Aptamer Assays (ELAA) [67,68]. ELAAs using aptamers as the capture and detection probe are often labelled with Horseradish Peroxidase (HRP) and have been shown to afford higher sensitivities than traditional ELISAs in the detection of a broad range of targets including mycotoxins [69]. Bonel et al. used magnetic beads functionalized with an OTA aptamer to detect an HRP-labeled OTA and free OTA in a competitive biosensor approach achieving detection limits of 70]. Based on the widespread commercial use of ELISAs in detection kits and homecare sensors, ELAA-based sensors with potential commercial applications and personal home-use assays have been reported. Gu et al. adapted the POC glucose biosensor model to design a personal glucometer device for the detection of OTA in wine samples [71]. They labelled a complementary DNA with the enzyme invertase, which in its bound state to the OTA-aptamer could not exhibit its catalytic activity. The invertase-labeled DNA was however released upon OTA target binding by the aptamer and was able to catalyze the hydrolysis of sucrose to glucose for signal readout.
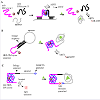
The activity of HRP-DNAzymes can be hindered by hairpin structures. The formation of hairpin structures with the target aptamer sequence locks the DNAzyme into a rigid stem-loop configuration such that the enzymatic function is efficiently blocked. Lee et al. designed a covalently conjugated hemin to a hairpin structure that loosened up in the presence of an OTA target (Figure 3B) [76], thus restoring the HRP-DNAzyme activity for a color change reaction. These studies all have a common signal-on approach where the enzymatic activity of the HRP-DNAzyme is lost initially and revived upon target introduction. In a different approach, Seok et al. described a signal-off variant by using split DNAzymes that forms a HRP-mimicking GQ structure on hybridization to an AFB1-aptamer and can therefore catalyze a color change reaction (Figure 3A) [77]. The aptamer preferentially binds to AFB1 on its introduction, thus releasing the split DNAzymes and resulting in the loss of its catalytic activity hence its signal readout. The idea that the HRP-DNAzyme aptamer constructs as described above can be used in a resonance energy transfer model has been explored. In this approach known as Chemiluminescence Resonance Energy Transfer (CRET), the HRP-DNAzyme acts either as a luminescence quencher, the luminescent species that is quenched, or an internal energy source for triggering luminescent species [78,79]. Jo et al. reported a signaloff CRET aptasensor based on dabcyl quenching to detect OTA in coffee samples (Figure 3C) [80]. They linked a dabcyl-labeled OTA aptamer with a HRP-DNAzyme GQ which catalyzed the oxidation f luminol for a light emitting signal. By introducing OTA, the OTAaptamer formed a GQ structure that brought the dabcyl quencher in close proximity to the DNAzyme thus quenching the signal.
GO surfaces can have heterogeneous adsorption affinities having sites with strong to moderate adsorption affinities. This affects the rate of aptamer adsorption/desorption from the GO surface. As aptamer sequences have to be desorbed from the GO surface to bind with targets, the turnover rate of targets introduced to signal generated is reduced, affecting the overall sensitivity of this platform. Liu et al. found that only one of six target DNA introduced in a GO sensor will generate a detectable signal. To reduce this effect [87], GO surfaces are usually modified for a homogenous size-specific adsorption and desorption using blocking agents such as polymers and surfactants which can increase its sensitivity by up to 10-fold. Zhang et al. tuned the size and dynamic range of a GO nano sheet and were able to eliminate non-specific adsorption in the fluorescence detection of AFB1 [88]. The sensor showed high sensitivity and selectivity for the AFB1 target in spiked food samples, and protected the AFB1 aptamer from nuclease denaturation.
Aptamer detections of metal ionsThe strong and selective binding of the DNA base, Thymine (T) and Hg2+ was first reported over a decade ago, and T-Hg2+-T interaction was used then to construct a sensor kit for Hg2+ detections [103]. The T-Hg2+-T bond results from the deprotonation and mercuration of the thymine N2, giving rise to N-Hg-N bond. This may distort the DNA structure, but strongly favours and stabilizes B-form DNA, having an extraordinary thermal stability owing to some form of metallophilic attraction of the Hg atom in the T-Hg2+-T base pair mismatch [104,105]. Aptamers are used in heavy metal detections due to their structure switching hybridizations with heavy metals. Aptamer sequences can also be easily obtained by in vitro selection as highly discriminatory binding species for heavy metal cations. The utility of aptamers in heavy metal detection is boosted by RNA-cleaving DNAzymes, and their ability in the presence of a divalent metal ion cofactor to cleave a substrate at its cleavage site thus introducing a signalling event that can be monitored with fluorophores, AuNPs, and other detection labels [106]. The HRPDNAzyme as discussed earlier is also used to detect metal ions and all these models will be discussed.
The catalytic activity of exonuclease III (ExoIII) in the specific stepwise cleavage of mononucleotides from duplex DNA has been exploited in Hg2+ detections. ExoIII will catalyse the stepwise removal of mononucleotides from 3’-terminus of duplex DNA. Despite the strong T-Hg2+-T duplex structure, ExoIII has been shown to catalyse the cleavage of T-Hg2+-T duplexes thus releasing the Hg2+ back into solution. Wang et al. reported a label free detection of Hg2+ using this approach [109]. They designed a hairpin structure with unpaired T-rich stem pairs which formed the T-Hg2+-T duplex on the introduction of Hg2+ (Figure 5C). In its unbounded state, ExoIII was incapable of nicking the 3′-strand of the hairpin structure, but could do so once the T-Hg2+-T duplex structure is formed on introduction of Hg2+. The release of the 3’-strand results in the formation of a GQ structure that binds with a GQ-specific IrIII complex which is luminescent and results in a switch-on emission signal. The advantages of most T-Hg2+-T detections is that in most cases Hg2+ is released back into solution during the signaling event, and a repetitive binding and cleaving can occur leading to signal amplifications and ultra-sensitive detections.
The 8-17 DNAzyme shows highly specific metal ion-dependent reactivity and catalysis and in the presence of its specific metal cofactor can cleave all chimeric RNA/DNA substrates. Furthermore, the 8-17 DNAzyme is easily obtained by SELEX and are thus assessable for use in the specific detection of heavy metals. In most detection models, the substrate RNA/DNA is appropriately labeled at one or opposite ends of its sequence such that the introduction of the specific metal ion allows the DNAzyme to cleave the substrate and separate the reporter label from its switch or quencher label generating either a signal-on or signal-off response. In one of these constructs a ferrocene-labeled DNA substrate was self-assembled on Au electrode for a redox-active signal which was lost when Pb2+ was introduced and induced the 8-17 enzyme strand to cleave the substrate for a signal-off response [120].
Aptamer Utility in Sensor Platformsfor the Detection of Toxins and Heavy Metals
Olasunkanmi O. Olaoye and Richard A. Manderville*
- Departments of Chemistry and Toxicology, University of Guelph, Canada
*Address for Correspondence: Richard A. Manderville, Departments of Chemistry and Toxicology, University of Guelph, Guelph, Ontario, N1G 2W1, Canada, Tel: (519)-824-4120; x53963; Fax: (519)-766-1499; E-mail: rmanderv@uoguelph.ca
Citation: Olaoye OO, Manderville RA. Aptamer Utility in Sensor Platforms for the Detection of Toxins and Heavy Metals. J Toxins. 2017;4(1): 12
Copyright © 2017 Olaoye OO, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Toxins | ISSN: 2328-1723 | Volume: 4, Issue: 1
Submission: 27 February, 2017 | Accepted: 14 March, 2017 | Published: 30 March, 2017
Abstract
Nucleic acid research has expanded in recent years through the development of an in vitro methodology for the derivation of functional nucleic acids, capable of binding to specific targets with high specificity and sensitivity. These activities are similar to those of antibodies, yet the single-stranded oligonucleotide, called aptamer, can be readily created on a large-scale using solid-phase oligonucleotide synthesis. This relative ease of synthesis has made aptamers a highly versatile tool for the detection of important targets for applications in imaging, diagnostics and therapeutics. Aptamers are able to show high fidelity to specific targets due to their selective affinities. They undergo structural alteration and topological changes when they bind to specific targets and most sensors using aptamers today leverage on this characteristic to record a detectable signal. The use of aptamers in modern applications is vast and cannot possibly be covered in a single review. This review explicitly focuses on DNA aptamers used for detecting toxins and heavy metals, and outlines their utility in contemporary sensor designs, and their future prospects as analytical tools.Keywords
Aptamers; Mycotoxins; Ochratoxin A; Aflatoxin; Fluorescence; DNAzymesIntroduction
Since their discovery by two independent research groups in 1990 [1,2], aptamers have become a versatile analytical detection tool used in research, diagnostics, and therapeutics. Aptamers are single-stranded oligonucleotides (ssDNA or ssRNA) that can adopt secondary and tertiary structures through target binding and changes in their environment. Aptamers are usually 15-80 bases long with affinities for a broad scope of biological and chemical targets ranging from large biomolecules such as proteins to smaller species such as metal ions. Aptamers are generated by SELEX [1,2] (Systemic Evolution of Ligands by Exponential Enrichment), which is an in vitro selection technology and recent advances to the SELEX process has afforded aptamers with diverse properties, capable of detecting a range of previously inaccessible targets [3-7]. Post-SELEX modifications have expanded the scope of aptamer applications in genomics [4] and proteomics [5], affording electroactive [8,9], colorimetric [10], and fluorescent properties for target elucidation [11,12].There have been several reviews in the past decade detailing aptamer applications in diagnostics and therapeutics [13-15], in sensor platforms [16-18], and in molecular imaging [19,20]. In this review the focus is placed on DNA aptamers designed to specifically detect food toxins and heavy metals. Environmental pollution has become a mainstay research interest in the past two decades as the invention and use of newer chemicals in agriculture, food processing, and consumer products have given rise to newer toxic substances that pose significant risks to living species. Food toxins such as mycotoxins (aflatoxins and ochratoxins) are widely studied and reviewed [21-24], and their persistence in food samples is a major global health concern. It is estimated that over 25% of crops produced are affected by mycotoxins [22], resulting in billions of dollars lost annually [24]. Based on their mechanistic mode of action [23,25,26], mycotoxins are implicated as carcinogenic, mutagenic, teratogenic, and tumorigenic. On the other hand, heavy metals are produced from various anthropogenic sources such as mining and solid waste combustion processes, and are released into soil and water bodies at levels disproportionate to their uptake by plants and microorganisms in soil and water habitats, and this poses significant health risks to the ecosystem. Heavy metals such as Cd, Ar, Cr, Hg, Pb, Ag, and Cu are known to be cytotoxic [27], carcinogenic and mutagenic [28-30], and exhibit toxicity even at low concentrations [30,31]. As heavy metals are non-biodegradable, they easily accumulate and persist in the environment polluting the food chain and disrupting the balance of the ecosystem [28]. The persistence of mycotoxins and heavy metals in the food chain is a major concern for food safety and consumer health, and viable removal mechanisms are being sought after.
Detection systems that can reveal the presence and identity of these pollutants in food systems are essential to guaranteeing food safety and this has become an objective of several research groups and laboratories. Mycotoxins such as aflatoxins and ochratoxins are currently detected by conventional laboratory chromatographic techniques such as Mass Spectrometry (MS), Gas Chromatography (GC), High Performance Liquid Chromatography (HPLC), and Thin Layer Chromatography (TLC). While these methods continue to offer accurate and highly sensitive detections, they are still plagued by an expensive and highly sophisticated instrumentation, a laboratorybased analysis that requires trained analysts, and the long hours required for sample collection, sample transfer to the laboratory, sample preparation and analysis, the entire process taking several days. These factors limit the applicability of chromatographic analysis as a rapid, easy to use, on-site detection tool. Immunoassays are an alternative to chromatography, but their limitations are in the use of pH and temperature sensitive antibodies, which makes them highly susceptible to denaturation, coupled with the challenges of growing antibodies in vitro and some toxin-specific antibodies in vivo [32-34].
The major analytical tools used in the detection of heavy metals are chromatographic (HPLC, MS, and GC), and spectroscopic (Atomic Absorption Spectroscopy (AAS) using flame, electro thermal, hydride, and non-dispersive protocols) [35,36]. Atomic Absorption Spectroscopy (AAS) is a fast, selective, and inexpensive analytical tool currently employed in the detection of heavy metals in food and water samples [35]. The current challenges of AAS are its high detection limits and a high influence of interfering matrix ions [35]. The high detection limit can be mitigated by pre-concentration steps using techniques such as extraction [37,38], filtration [39], and electrode position to amplify the metal ion analyte signal [40].
The analytical detection methods outlined above do not satisfy the current food industry demands for an easy to use, on-site molecular imaging detection tool required for ultrasensitive determination of heavy metals and toxins in food samples. More recent research focus has thus shifted to the development of DNA aptamer biosensors as a feasible alternative in detecting food contaminants at very low concentrations. Aptamers are strong favorites to herald in cheap, easy-to-use, and highly sensitive analytical devices that can ensure food safety due to their following properties: (1) aptamers have high binding affinity and specificity for their specific targets, (2) aptamers undergo conformational changes upon ligand binding adopting secondary and tertiary structures, an event that is easily monitored by several detection means [41], (3) aptamers are stable and highly versatile for incorporation into various transduction platforms (electrochemical, mechanical, and optical detections), making them suitable for integration into different designs and use in miniaturized devices [18], (4) post-SELEX modifications afford tailored properties such as stability, fluorescence, denaturation resistance, enhanced selectivity and affinity depending on the final binding conditions desired [7], (5) they undergo Polymerase Chain Reactions (PCR), which can be used to synthesize the DNA aptamer sequence to produce more copies, and enhance transduction signals especially when detecting trace targets [42], and (6) aptamers can be developed for in vivo applications, as they are neither carcinogenicnor immunogenic, and will elicit no undesired cellular response. Thus, over the past two decades, DNA aptamer biosensors have been in rapid development and useful platforms have emerged for sensing toxins and metals, suggesting a strong future for DNA aptamers as tools for ensuring food and water safety.
Aptamer detections of mycotoxins
Single fluorophore systems: The basic technique used to generate probe aptamers is to covalently attach an extrinsic fluorophore to the aptamer nucleobase where a conformational change is expected to occur upon target binding (Figure 1A, (I)). This approach relies upon a conformational change actually occurring at the attached fluorophore site, that is accompanied by an alteration of the fluorophore groups electronics that can elicit a change in its intrinsic fluorescent properties [43,44]. This monochromorphic approach was used by Zhao et al. in a Fluorescein Amidite (FAM) labeled aptamer to detect Ochratoxin A (OTA) [45]. The FAM dye was labeled at the 3′, 5′, and the Thymine (T) sites of the aptamer, and conformational changes during OTA binding induced a fluorescent turn-on or turn-off response by the attached fluorophore emission. Due to the uncertainties of this approach, several logically designed fluorophore-labeled constructs have to be tested in order to obtain the desired signalling aptamer. Jhaveri et al. had to test several aptamer constructs, inserting fluorophore labels at seven different locations along the aptamer sequence to identify the most suitable fluorophoreinsertion site [44]. Only two of their seven modified aptamers gave a fluorescent signal upon target binding, and the increase observed in fluorescent intensities was <50% in both cases. To date, single fluorophore systems have been shown to be unsuitable as models for generic aptasensor (aptamer sensor) designs and exhibit lower sensitivities compared to other models.Aptamers that can switch from a complementary DNA-aptamer duplex to a tertiary target-aptamer structure are used in various designs to detect small molecules and proteins. These aptamers can be generated by a structure-switching SELEX selection process that directly produces the required aptamer sequence [41,51]. These constructs are designed to have the fluorophore or quencher on either of the aptamer or the complementary DNA sequence such that when the aptamer binds to the target, there is definitely a separation of the fluorophore and quencher which is not dependent on an accurate knowledge of the aptamer-target tertiary structure (Figure 1C). This approach was used by Chen et al. to detect trace amounts of OTA in corn samples at detection levels 52]. A fluoresceinlabeled OTA aptamer was made to hybridize with a quencher-labeled complementary DNA which effectively quenched the emission of the labeled fluorophore. On introduction of corn samples spiked with OTA, the aptamer unbounds from the DNA duplex to form an aptamer-OTA duplex, thus reviving the fluorescence of the attached dye. Structure-switching FRET aptasensor constructs adopting this approach show high sensitivity and selectivity, and have very low detection limits for their targets [53].
Fluorescent nucleobase approaches: Fluorescent dyes are widely used as aptamer labels to report structural changes due to their high quantum yields and molar absorptivity. These dyes are commercially accessible and easily incorporated into oligonucleotide structures [54]. However, fluorescent dyes are bulky groups that can introduce steric hindrance, upset the natural structure of the nucleic acid, inhibit nucleobase interactions and weaken biological activities. These structural limitations require that fluorescent dyes be inserted via flexible, long alkyl chain linkers which makes their orientation and position in the DNA structure highly unpredictable [55,56]. As natural nucleobases are intrinsically non-emissive, modified nucleobase probes that can retain a high fluorescence yield has become an interesting area of aptamer research [56,57]. Unlike fluorescent dyes, modified fluorescent nucleobases interact closely with their nearby bases through π-stacking and hydrophobic interactions. These modifications are small, rigid and may not largely alter the native aptamer structure, making them retain their biological activity and chemical interactions. They are also more suited to report changes in their environment due to the preciseness in their location and orientation [58].
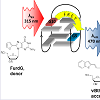
Despite the increased number of nucleobase-modified aptamers that have been reported as target-signaling probes [55-61], none have been successfully applied in the detection of mycotoxins. Despite the advantages these modified aptamers offer, their chemistry is still far from fully developed. Most modified fluorescent nucleobases require extending their π conjugation or introducing Donor-Acceptor (D-A) groups which alters the nucleobase structural preference and may distort base stacking interactions. These analogues can lose or possess weakened binding interactions for target structures, thus becoming unsuitable for some applications. Recent studies by the Manderville laboratory, however, strongly suggest that aptamers with modified fluorescent nucleobases for the detection of mycotoxins will soon become a reality [62-64]. The incorporation of fluorophore groups at the 8-position of purines is a useful way of reporting topological changes in structural conformation and G-Quadruplex (GQ) folding of nucleic acids [11,12]. The Manderville laboratory has incorporated isomorphic, visibly emissive 8-aryl-2′-deoxyguanosine (8-Ar-dG) probes into the Thrombin Binding Aptamer (TBA) to monitor GQ-folding and thrombin binding to the antiparallel GQ produced by TBA [12,59,62-64]. The 8-Ar-dG probes typically favour the syn-conformation, and this highly stabilizes the antiparallel GQ structure produced by TBA when placed in the G-tetrad at a syn-G site. Furthermore, the emission intensity of 8-aryl-dG bases, such as 8-furyl-dG (FurdG) exhibits quenched emission in the duplex that lights-up in the GQ structure, providing a useful turn-on signal for monitoring duplex-GQ exchange. By introducing a vinyl linker to connect the 8-aryl group to the dG component, a higher degree of rotation is permitted that allows favourable anti-conformational insertion, thus permitting Watson-Crick (W-C) base pairing of duplex DNA without perturbing GQ formation. With the vinyl linker, 8-Ar-dG probes can be excited at ∼ 390 nm, which is ostensibly far removed from unmodified nucleobase excitation, and this excitation overlaps with the excitation maxima of other 8-Ar-dG probes, such as FurdG. With this idea in mind, a FRET-based aptamer probe was designed by incorporating FurdG and 8-vinylbenzo[b]thienyl-dG (vBthdG) at different syn-positions (i.e G10 and G5) of TBA (Figure 2) [62]. The emission of the FurdG donor at ∼ 390 nm overlaps with the absorption maxima of the vBthdG acceptor, which provides a visible blue emission at ∼ 470 nm. The vBthdG probe alone was not effective as a single structure-switching probe for monitoring duplex-GQ exchange, as its emission is not adequately sensitive to DNA structural changes. However, in the FRET-based approach (Figure 2), the FurdG can act as a fluorescent switch turning on the visible emission of the vBthdG probe. These studies highlighted that 8-Ar-dG probes have emissive properties that enable them to act as fluorescent reporters of structure-switching aptamers during target binding, and an approach based on this model for the detection of mycotoxins is currently in development.
The obvious challenge with linking enzymes to aptamers or small molecules is the time consuming protocol required and the possible denaturation of the enzyme over time based on storage conditions. The knowledge that nucleic acid sequences with enzymelike catalytic activity can be obtained by in vitro selection has birthed the DNAzyme, which can be used in place of protein enzymes in ELAA-based assays. Unlike protein enzymes, DNAzymes are stable and can retain their enzymatic activity under different conditions of pH and temperature, are easily attached to their recognition sites through DNA base pairing interactions, can be easily identified and selected for specific targets by SELEX, and are easily synthesizedand multiplied by PCR [72]. The HRP-mimicking DNAzyme (HRP-DNAzyme) has been investigated as a reporter of aptamer interactions with small molecules, heavy metals, and proteins for colorimetric and chemiluminescence detections [73]. The HRPDNAzyme effectively catalyzes the oxidation of ABTS2- (2,2′-azinobis[ 3-ethylbenzthiazoline-6-sulfonic acid]) and luminol both in the presence of H2O2 for colorimetric and chemiluminescent detections (Figure 3). These detections have the value of not requiring an external light source such as in fluorescent detections as they are broughtabout by enzymatic/chemical redox reactions that leads to a change in the electronic properties of the sensing species. Shim et al. conjugated an HRP-DNAzyme with an Aflatoxin B1 aptamer (AFB1-aptamer) which effectively quenched the enzymatic property of the DNAzyme in the hybridized state [74]. On binding to AFB1, the DNAzyme was partially freed in its partially hybridized state and formed a hemininduced GQ structure allowing restoration of its HRP-mimicking catalysis for a visible emission signal. In a similar approach, Yang et al. used an obstructive oligonucleotide sequence that partially hybridizes to both an OTA-aptamer and the HRP-DNAzyme thus inactivating the DNAzyme [75]. The presence of OTA loosened the obstruction, and in the presence of K+ and hemin, the DNAzyme forms the HRPmimicking GQ structure for a colorimetric detection.
Figure 3: Various configurations of DNAzymes-aptamer interactions for mycotoxin detection, (A) signal-off split DNAzymes that lose HRP mimicking property upon target binding, (B) signal-on aptamer DNAzyme hairpin probe, and (C) CRET-based signal-off aptamer-DNAzyme probe based on dabcylquenching upon target binding.
A notable benefit of the HRP-mimicking DNAzymes is that their signal can be readily amplified up to 1000-fold by various phenolic substrates, a feature that makes their detections highly sensitive [81]. They also show versatility in various aptasensor constructs based on the amenability of their nucleic acid structure and the functional applications this can allow such as in PCR and Hybridization Chain Reactions (HCR). Wang et al. utilized HCR to offer an ultra-sensitive colorimetric detection of OTA in wine and wheat samples [82]. They designed separate aptamer and HRP-DNAzyme hairpin structures with complementary 5′ and 3′ sequences and a central aptamer sequence and central DNAzyme sequence respectively. The aptamer hairpin opens up upon OTA binding allowing its 3′ sequence to hybridize with the complementary 3′-end sequence of the DNAzyme. This opens up the DNAzyme hairpin to form an HRP-DNAzyme GQ structure that catalyzes the color change oxidation reaction of ABTS. As the 5′-end sequence of both structures are also complementary, there was repetitive aptamer binding and opening up that led to HCR and afforded enhanced detection of OTA. These studies demonstrated the utility of aptamers as adaptable, structure-switching probes, their design possibilities that stretches as far as our creativity and promises cheaper and simpler detection platforms for target molecules.
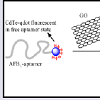
Aptamer-Graphene interactions: Graphene has received enormous research attention due to its extraordinary properties of conductivity, electronics, surface area, tunability, and core strength. Graphene is a carbon nanomaterial that has been shown to form Π-Π stacking interactions with DNA bases which affords strong adsorption of DNA to its 2D monolayer [83]. Graphene has attracted significant interest and use in aptamer probe designs due to its preferential adsorption of only ssDNA and its ability to quench the emission of fluorescent species. Hence fluorescent labeled-ssDNA can be desorbed from its surface by target binding or to a complementary DNA sequence for signal-on detections. The structural adsorption preference of graphene was exploited in the SELEX-screening and selection of novel aptamers for the detection of the mycotoxin, patulin [84].Carbon Nanotubes (CNTs), Graphene Oxide (GO) and reduced GO (rGO) are the variants of graphene that are reported in most aptamer probe devices due to their high metal-free purity, and their very cheap mass manufacture process. Driven by a massive conjugated structure, GO possesses an intrinsic energy/electron transfer character over a broad wavelength range, allowing it to act as a possible quencher for suitable fluorescent probes. GOs can act as excellent electronic DA pair, quenching the fluorescence of several emissive species such as organic dyes, NPs, and Quantum Dots (QD). Lu et al. designed a Cadmium-telluride QD (CdTe- QD) modified AFB1-aptamer that exhibited a CdTe-QD - GO fluorescence quenching in the absence of the AFB1 target due to the strong adsorption of the aptamer to the GO surface (Figure 4) [85]. On introduction of the AFB1 target, the AFB1 aptamer detaches from the GO-surface and binds to AFB1 to form a GQ structure, effectively restoring the fluorescence of CdTe-QD. CNTs and rGOs employ similar fluorescence quenching mechanism when used to monitor structure-switching detections. Lv et al. described an OTA-aptamer sensor based on the fluorescence quenching of an attached FAM label by single-walled CNT, and the restoration of its fluorescence upon OTA target binding [86].
GO surfaces can have heterogeneous adsorption affinities having sites with strong to moderate adsorption affinities. This affects the rate of aptamer adsorption/desorption from the GO surface. As aptamer sequences have to be desorbed from the GO surface to bind with targets, the turnover rate of targets introduced to signal generated is reduced, affecting the overall sensitivity of this platform. Liu et al. found that only one of six target DNA introduced in a GO sensor will generate a detectable signal. To reduce this effect [87], GO surfaces are usually modified for a homogenous size-specific adsorption and desorption using blocking agents such as polymers and surfactants which can increase its sensitivity by up to 10-fold. Zhang et al. tuned the size and dynamic range of a GO nano sheet and were able to eliminate non-specific adsorption in the fluorescence detection of AFB1 [88]. The sensor showed high sensitivity and selectivity for the AFB1 target in spiked food samples, and protected the AFB1 aptamer from nuclease denaturation.
Other designs using aptamers for mycotoxin detections: While there are several aptamer models for mycotoxin detection, only very few can be covered in one review. The vast potential of aptamer integration into various sensing and transduction platforms has led to the increase in aptasensor design possibilities. One of these possibilities reported by Zhang et al. uses Terbium (Tb3+), a rare earth metal ion, and exploits the ability of ssDNA to amplify the fluorescence emissions of Tb3+ in solution by magnetic separation [89]. By hybridizing two ssDNA with an OTA aptamer to form a GQ structure, they effectively quenched the fluorescence observed from Tb3+ in solution. The OTA aptamer however detaches from the ssDNAs to bind with OTA when it is present, freeing up the ssDNA to return to solution thus reviving the fluorescence observed from Tb3+. Based on the ELAA model, Costantini et al. reported a low-cost microfluidic chip aptasensor based on an array of amorphous silicon photosensors for a highly sensitive detection of OTA in beer samples. In a very different approach [90], Liu et al. designed a cross-linked aptamer hydrogel to detect OTA [91]. The hydrogel network was formed by hybridizing an OTA-aptamer with two complementary DNA strands containing loaded AuNPs. Aptamer binding with the OTA target led to hydrogel dissociation that permitted the release of the preloaded AuNPs. The release of AuNPs can be observed by the naked eye for a visible detection, and the introduction of an Au@Pt core-shell NP driven Volumetric bar-chart Chip (V-Chip) enabled quantification of the OTA present. Provides a list of different models and probe designs that further highlight the utility and versatility of aptamers in mycotoxin sensor designs (Table 1) [92-102].
Aptamer detections of metal ionsThe strong and selective binding of the DNA base, Thymine (T) and Hg2+ was first reported over a decade ago, and T-Hg2+-T interaction was used then to construct a sensor kit for Hg2+ detections [103]. The T-Hg2+-T bond results from the deprotonation and mercuration of the thymine N2, giving rise to N-Hg-N bond. This may distort the DNA structure, but strongly favours and stabilizes B-form DNA, having an extraordinary thermal stability owing to some form of metallophilic attraction of the Hg atom in the T-Hg2+-T base pair mismatch [104,105]. Aptamers are used in heavy metal detections due to their structure switching hybridizations with heavy metals. Aptamer sequences can also be easily obtained by in vitro selection as highly discriminatory binding species for heavy metal cations. The utility of aptamers in heavy metal detection is boosted by RNA-cleaving DNAzymes, and their ability in the presence of a divalent metal ion cofactor to cleave a substrate at its cleavage site thus introducing a signalling event that can be monitored with fluorophores, AuNPs, and other detection labels [106]. The HRPDNAzyme as discussed earlier is also used to detect metal ions and all these models will be discussed.
T-Hg2+-T Interactions: Aptamers with T-T mismatches can be functionally labeled to report the T-Hg2+-T binding event, and this has been the basis for constructing most aptamer models for Hg2+ detections. The molecular beacon construct is a widely used model in detecting nucleobase-Hg interactions, as a conjugate stem sequence can be functionalized with a suitable label such that it can report changes in the molecular beacon structure during Hg2+ binding. Wang et al. attached a fluorophore to the 5′-end of a T-rich hairpinforming sequence, and a quencher to a partially complementary sequence that disrupted the formation of the hairpin structure [107]. The introduction of Hg2+ assisted the release of the complementary sequence and formation of the hairpin structure that allowed the fluorophore to interact with an evanescent receptor molecule producing an evanescent wave induced fluorescent detection (Figure 5A). To further exploit the strong T-Hg2+-T interaction andthe structure switching properties of aptamers, Li et al. designed a gate-controlled electrochemical probe for Hg2+ detection (Figure 5B) [108]. Their strategy involved a SH-β-Cyclodextrin (β-CD) assembled Au-electrode on which a thionine-labeled Hg2+-specific aptamer-complementary DNA sequence duplex structure was linked. The introduction of Hg2+ detaches the aptamer sequence from the complementary DNA to form a hairpin structure bringing the thionine label in close proximity with Au electrode and thus blocking the interstitial spaces of the β-CD which served as channels for an electrochemical probe. Thus the probe could not gain access to the Au electrode resulting in a decrease in the probes oxidative current.
Figure 5: Different aptamer constructs for heavy metal detection, (A) hairpin-forming T-Hg2+-T binding cleaves complementary DNA, (B) gatecontrolled electrochemical probes based on Hg2+-induced aptamer folding, (C) exonuclease assisted cleavage of T-Hg2+-T hairpin to form a GQ HRPDNAzyme, and (D) Cd2+-induced hirpin formation for signal-on fluorescence.
In vitro selection: The selection process for aptamers that show highly selective and strong binding affinity for metal ions could have its challenges. For example, small molecules such as metal ions tend to complicate the separation process during aptamer selection as there is no distinct difference in the size of free and bound aptamers. This generally leads to a poorer aptamer selection process, giving rise to a final sequence with a reduced dissociation constant (Kd) for the target. To reduce this limitation, several variations of SELEX immobilizes the oligonucleotide library rather than the target species [110]. A second challenge is in the specificity of the selected aptamer sequence for metal ions, as most metal ions with similar sizes and valencies usually have similar characteristics and may be undistinguishable by the aptamer sequence. One way of reducing this effect is by introducing negative selection rounds to eliminate potential promiscuous aptamer sequences. Based on these two limitations, Wang et al. reported a SELEX process modified to identify a highly selective aptamer sequence for Cd2+ detection (Figure 5D) [111]. In their approach, a biotin-labeled aptamer-complementary oligonucleotide library was effectively immobilized on streptavidin-agarose beads, and the introduction of Cd2+ detached the sequences with strong affinity for Cd2+. A round of negative SELEX was introduced to isolate sequences with affinity for Zn2+, which is the closest divalent metal to Cd2+. This selection process resulted in a structure-switching hairpin aptamer that showed strong affinity and preferential selectivity for Cd2+ against other metal ions in a FRET detection model.
Graphene and NPs: Fluorescent detections of metal ions using graphene in a D-A electron/energy transfer model as discussed earlier has been reported. Qian et al. reported a QD-labeled aptamer whose fluorescence was quenched when immobilized on GO by photo-induced electron transfer [112]. The introduction of Pb2+ for preferential binding of the aptamer and its simultaneous detachment from the GO surface resulting in the fluorescence recovery of the emissive graphene QD. The ease of aptamer integration to adsorption surfaces such as graphene and NPs has afforded sensing devices of varying detection protocols. Cui et al. described a SPCE-AuNP selfassembled As3+ aptamer that was capable of repelling other cationic species by its adsorption of polydiallyldimethylammonium (PDDA) [113]. However, the presence of As3+ changes its conformation from hairpin to GQ, and preferentially adsorbs the more cationic [Ru(NH3)6]3+ which acts as the electrochemical indicator to produce a signal-on response. Zhang et al. used T-rich or G-rich DNA molecular beacons bounded to Digital Video Discs (DVD) for the detection of Hg2+ or Pb2+ respectively [114]. The hairpin structure opened either by T-Hg2+-T interaction or the formation of GQ allowing a bounded biotin interact with the AuNP-streptavidin conjugate for a quantifiable digital signal that was processed by an optical drive.
DNAzymes in the detection of heavy metals: The extraordinary stability and preferential binding of the T-Hg2+-T interaction has also been exploited in an HRP-DNAzyme construct for the detection of Hg2+. HRP-DNAzymes in their GQ structures possesses enzymatic activity when Hg2+ is absent, but preferentially unfolds from the GQ structure when Hg2+ is introduced losing their peroxidase catalytic activity. Tang et al. reported a dual detection of Hg2+ and Metallothionenes (MTs) based on this approach [115]. MTs have a strong capacity to bind heavy metal cations with the thiol group of their cysteine residue. The HRP-DNAzyme lost its catalytic activity when it unfolds from its GQ structure to form the T-Hg2+-T base pair mismatch but the introduction of MTs lead to a MT-Hg2+ interaction and restoration of the DNAzymes GQ structure. The DNAzyme can hence catalyse the color change oxidation of ABTS2- for signal readout.HRP-DNAzymes have been used in the detection of heavy metals other than Hg2+, especially metal cations that alter the character of a K+-stabilized GQ. The fact that Pb2+ can strongly stabilize a GQ structure has been exploited, and the character of a Pb2+-stabilized GQ has been studied extensively in order to design an effective probe based on this approach. It has been shown that Pb2+ induces a unimolecular folded GQ with TBA and a conformational switch in the PS2.M GQ when it displaces K+ and Na+ from its GQ planes and central cavities respectively [116]. The PS2.M GQ is of special interest as it shows superior peroxidase activity when complexed with hemin and this is the basis of PS2.M detection of Pb2+. In one study, Wang et al. found that when Pb2+ displaces K+ from the PS2.M GQ plane, the consequent conformational switch results in loss of catalytic activity and this was measured by loss of luminescence in a PS2.M catalyzed H2O2-mediated oxidation reaction of luminal [117]. Most studies that utilize PS2.M to detect Pb2+ rely on the loss or diminished catalytic activity of PS2.M when Pb2+ displaces K+ from its binding planes. In a different approach however, Liu et al. found that Pb2+ can induce GQ formation of a G-rich DNAzyme that shows peroxidase activity and catalyzes the color change of H2O2-mediated oxidation of a guaiacol indicator [118]. The DNAzyme in the presence of Pb2+ unimolecularly folds into the HRP-DNAzyme GQ that catalyzes the oxidation of colorless guaiacol into the amber tetraguaiacol, and this concept was extended into a detection kit for Pb2+ in real food samples.
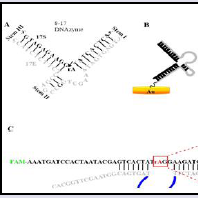
DNAzymes that catalyze the cleavage of RNA bonds in the presence of metal ions were first selected by in-vitro selection in 1994. Since then, several RNA-cleaving DNAzymes have been identified including the two most studied 8-17 and 10-23 DNAzymes [119]. The 8-17 and 10-23 DNAzymes are capable of cleaving all RNA substrates at their phosphodiester linkage in the presence of a divalent metal ion cofactor. The metal ion is required not only for its electronic activity but also plays a structural role. The 8-17 DNAzyme has an all DNA oligonucleotide enzyme strand (17E) and a chimeric DNA/RNA substrate strand (17S) which has a cleavable riboadenosine (rA) site (Figure 6A). The 17E binds 17S by W-C base pairing to form 3 hairpin stems looped together by 5 unpaired nucleobases.
In expanding the utility and structure switching properties of DNA aptamers, Zhou et al. designed an electrochemical sensor based on Pb2+-induced 8-17 DNAzyme catalysis, HRP-DNAzyme GQ, and Au-Pd bimetallic NPs for the detection of Pb2+ (Figure 6B) [121]. They immobilized the 8-17 DNAzyme on Au electrode and in the presence of Pb2+ the substrate DNA was cleaved exposing the enzyme strand which remained immobilized. Simultaneously, several strands of a GQ forming G-rich DNAzyme sequence were immobilized on Au-Pd NP. One of this G-rich HRP-DNAzyme strand is enabled to hybridize with the exposed enzyme strand of the 8-17 DNAzyme with the aid of a helper DNA sequence. The addition of hemin leads to the formation of HRP-DNAzyme GQ for all the unhybridized G-rich DNazyme strands, and the formed HRP-DNAzyme GQs catalyzes the reduction of H2+O2+ in the presence of thionine as the electron mediator to generate an electrochemical signal.
Modifying the RNA cleaving site of substrate-cleaving DNAzymes has led to new possibilities in heavy metal sensing. Wang et al. replaced the cleavable rA site of the 8-17 DNAzyme to an uncleavable rA [122], whilst retaining its ability to report heavy metal binding as shown by their graphene Field Effect Transistor (FET)-based detection of Pb2+. Liu and Huang were able to induce Cd2+ specificity for a RNA-cleaving DNAzyme by modifying the phosphate (P-O bond) RNA linkage to a phosphorothioate bond (P-S) which allowed more reactivity with thiophilic metals such as Cd2+ (Figure 6C) [124]. By a careful in vitro selection process, and introduction of negative selection rounds to exclude sequences with affinity for similar thiophilic metals such as Pb2+ and Hg2+, a DNAzyme was selected and shown to have exceptionally specific Cd2+-dependent activity and was employed in the selective detection of Cd2+ in rice samples.
As with many detection platforms that employ aptamers and nucleic acid structures as recognition and detection sensors, the detection of heavy metals with RNA cleaving-DNAzymes shows remarkable utility and versatility, and as such several models have been reported. Tsekenis et al. designed an RNA-cleaving DNAzyme formed by hybridizing a laser printed and surface sensor-immobilized 17-E strand with the 17-S DNA substrate which was cleaved on introduction of Pb2+ to induce a surface stress measured by changes in the device capacitance [124]. Zhang et al. reported a Pb2+ sensor based on the personal glucometer [125]. They immobilized the RNAcleaving DNAzyme on a microplate that contains invertase-labeled ssDNA strands immobilized on AuNPs. The enzyme-labeled DNA strands are complementary to the DNAzyme substrate strand and hybridization occurs through a Pb2+-induced substrate cleavage leading to a signal read-out. Li et al. exploited GO-ssDNA Π- Π interactions [126]. They modified the 8-17 DNAzyme substrate with AuNPs thus restricting any DNAzyme interaction with GO, but thepresence of Pb2+ cleaved the substrate strand, allowed the cleaved ssDNA to be adsorbed on the GO surface and the AuNPs to interact with the GO for a colorimetric signal-on detection.
Future Directions
Though aptamers continue to show promise as the next generation diagnostic and sensing probes, their application in real life devices have been significantly limited despite the high-level research interest it has attracted over the last decade. To present, very few aptamers have any commercial value, and aptamers for several targets are yet to be identified, or at best are still at a proof-of-concept stage. Aptamer commercialization competes against a well-developed antibodybased therapeutic and diagnostic industry, and replacing antibodies will take its own time considering the huge investments that was required before antibody technology came to fruition. Aptamers are thus increasingly being explored for Laboratory Developed Tests (LDTs) in food safety and environmental analytes such as NeoVentures Biotechnologies affinity columns and ELAA kits for OTA and aflatoxins. The commercialization of aptamer detection kits for food toxins may just be on its way based on recent ultra-sensitive and rapid aptamer assays that is able to probe large quantities of food cutting off days from traditional ELISAs′ culture enrichment and assay times [14,52,69,102]. New aptamer base analogues also expand the reporting adaptability of aptamers beyond their four nucleobase systems, and these unnatural analogues offer proven advantages in assay performance and stabilizing aptamer structures against pH, temperature, salt and ionic concentrations, and other denaturation conditions. It is expected that modified nucleobases will continue to be explored, and a viable modified aptamer will soon be available providing the shift to cheaper, faster, highly sensitive and selective detection kits for mycotoxins and heavy metals.The selection of aptamers for small molecules continues to have its own challenges [127]. It has been noted that aptamers for small molecules such as mycotoxins and heavy metals have lower affinities when compared to aptamers for larger molecules such as proteins. This is mainly attributed to the reduced target size, immobilization protocols, which are painstaking for smaller molecules, and the almost constant presence of interfering species. The selection process also is not well defined, and no ideal generic protocol is currently used. Depending on the applicability of the aptamer been sought, researchers have to choose from a range of selection models with each having its own limitation of cost, time, equipment, and availability of the needed oligonucleotide library. Advances in robotic and in-silico SELEX applications offer the hope of a one-step selection process that is expected to streamline and simplify aptamer identification, and an ideal selection tool for future aptamer discoveries.
Conclusion
This review explored recent innovations in sensing devices for mycotoxins and heavy metals that highlights the utility of aptamers as structure switching signalling probes. Current understanding of aptamer structure and target interactions provides a highly reliable sensing platform that rivals antibody-based detections. The commercial applicability of aptamer kits is expected to improve over the coming years, especially with improvements in the selection and post-identification process of aptamer sequences. Aptamer based assay kits are expected to become a strong choice in the future detections of food toxins and heavy metals. Overall, aptamers offer a very promising approach to new generation detection protocols for mycotoxin and heavy metals.Acknowledgements
Support for this research was provided by the Natural Sciences and Engineering Research Council (NSERC) of Canada, the Canada Foundation for Innovation, the Ontario Innovation Trust Fund.References
- Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment - RNA ligands to bacteriophage-T4 DNA-polymerase. Science 249: 505-510.
- Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818-822.
- Yuce M, Ullah N, Budak H (2015) Trends in aptamer selection methods and applications. Analyst 140: 5379-5399.
- Ruff P, Storici F (2016) Genome editing by aptamer-guided gene targeting (AGT). In: Cathomen T, Hirsch M, Porteus M (Eds). Genome editing: The next step in gene therapy. Springer, New York, pp. 111-124.
- Ma H, Liu J, Ali M, Mahmood M, Labanieh L, et al. (2015) Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem Soc Rev 44: 1240-1256.
- Blind M, Blank M (2015) Aptamer selection technology and recent advances. Mol Ther Nucleic Acids 4: e223.
- Darmostuk M, Rimpelova S, Gbelcova H, Ruml T (2015) Current approaches in SELEX: An update to aptamer selection technology. Biotechnol Adv 33: 1141-1161.
- Xiang Y, Xie M, Bash R, Chen J, Wang J (2007) Ultrasensitive label-free aptamer-based electronic detection. Angew Chem-Int Ed Engl 46: 9054-9056.
- Ferapontova E, Gothelf K (2011) Recent advances in electrochemical aptamer-based sensors. Current Org Chem 15: 498-505.
- Rhouati A, Yang C, Hayat A, Marty J (2013) Aptamers: a promising tool for ochratoxin a detection in food analysis. Toxins 5: 1988-2008.
- Sproviero M, Fadock KL, Witham Aaron A, Manderville RA (2015) Positional impact of fluorescently modified G-tetrads within polymorphic human telomeric G-quadruplex structures. ACS Chem Biol 10: 1311-1318.
- Gomes A, Das R, Sarkhel S, Mishra R, Mukherjee S, et al. (2010) Herbs and herbal constituents active against snake bite. Indian J Exp Biol 48: 865-878.
- Xiang D, Shigdar S, Qiao G, Wang T, Kouzani A, et al. (2015) Nucleic acid aptamer-guided cancer therapeutics and diagnostics: the next generation of cancer medicine. Theranostics 5: 23-42.
- Bruno J (2015) Predicting the uncertain future of aptamer-based diagnostics and therapeutics. Molecules 20: 6866-6887.
- Famulok M, Hartig J, Mayer G (2007) Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev 107: 3715-3743.
- Famulok M, Mayer G (2011) Aptamer modules as sensors and detectors. Acc Chem Res 44: 1349-1358.
- Cho EJ, Lee JW, Ellington AD (2009) Applications of aptamers as sensors. Annu Rev Anal Chem (Palo Alto Calif) 2: 241-264.
- Song S, Wang L, Li J, Zhao J, Fan C (2008) Aptamer-based biosensors. Trends in Anal Chem 27: 108-117.
- Jin C, Zheng J, Li C, Qiu L, Zhang X, et al. (2015) Aptamers selected by cell-SELEX for molecular imaging. J Mol Evol 81: 162-171.
- Hong H, Goel S, Zhang Y, Cai W (2011) Molecular imaging with nucleic acid aptamers. Curr Med Chem 18: 4195-4205.
- Bryden WL (2007) Mycotoxins in the food chain: human health implications. Asia Pac J Clin Nutr 16 Suppl 1: 95-101.
- Bhat R, Rai R, Karim A (2010) Mycotoxins in food and feed: present status and future concerns. Comp Rev Food Sci Food Safety 9: 57-81.
- Hussein HS, Brasel JM (2001) Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology 167: 101-134.
- Pfohl-Leszkowicz A, Manderville RA (2007) Ochratoxin A: an overview on toxicity and carcinogenicity in animals and humans. Mol Nutr Food Res 51: 61-99.
- Ronald R (1998) Mechanistic interactions of mycotoxins: theoretical considerations. In: Shinha KK, Bhatnagar D, (Eds). Mycotoxins in agriculture and food safety. New York: Marcel Dekker Inc; pp. 135-227.
- Speijers GJ, Speijers MH (2004) Combined toxic effects of mycotoxins. Toxicol Lett 153: 91-98.
- Morcillo P, Esteban MA, Cuesta A (2016) Heavy metals produce toxicity, oxidative stress and apoptosis in the marine teleost fish SAF-1 cell line. Chemosphere 144: 225-233.
- Ali H, Khan E, Sajad MA (2013) Phytoremediation of heavy metals--concepts and applications. Chemosphere 91: 869-881.
- Wu X, Cobbina SJ, Mao G, Xu H, Zhang Z, et al. (2016) A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environ Sci Pollut Res Int 23: 8244-8259.
- Skaar DA, Murphy SK, Hoyo C (2016) Effects of environmentally acquired heavy metals and nutrients on the epigenome and phenotype. In: Hughes CL, Walter MD, (Eds). Translational toxicology: Springer International Publishing, pp. 139-170.
- Dixit R, Wasiullah, Malaviya D, Pandiyan K, Singh UB, et al. (2015) Bioremediation of heavy metals from soil and aquatic environment: an overview of principles and criteria of fundamental processes. Sustainability 7: 2189-2212.
- Tombelli S, Minunni M, Mascini M (2007) Aptamers-based assays for diagnostics, environmental and food analysis. Biomol Eng 24: 191-200.
- Tombelli S, Minunni A, Mascini A (2005) Analytical applications of aptamers. Biosens Bioelectron 20: 2424-2434.
- Jayasena SD (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45: 1628-1650.
- Mendil D, Karatas M, Tuzen M (2015) Separation and preconcentration of Cu(II), Pb(II), Zn(II), Fe(III) and Cr(III) ions with coprecipitation method without carrier element and their determination in food and water samples. Food Chem 177: 320-324.
- Jain Ck, Ali I (2000) Arsenic: occurrence, toxicity and speciation techniques. Water Res 34: 4304-4312.
- Khazaeli S, Nezamabadi N, Rabani M, Panahi HA (2013) A new functionalized resin and its application in flame atomic absorption spectrophotometric determination of trace amounts of heavy metal ions after solid phase extraction in water samples. Microchem J 106: 147-153.
- Mashhadizadeh MH, Amoli-Diva M, Shapouri MR, Afruzi H (2014) Solid phase extraction of trace amounts of silver, cadmium, copper, mercury, and lead in various food samples based on ethylene glycol bis-mercaptoacetate modified 3-(trimethoxysilyl)-1-propanethiol coated Fe3O4 nanoparticles. Food Chem 151: 300-305.
- Alothman ZA, Unsal YE, Habila M, Tuzen M, Soylak M (2015) A membrane filtration procedure for the enrichment, separation, and flame atomic absorption spectrometric determinations of some metals in water, hair, urine, and fish samples. Desalin Water Treat 53: 3457-3465.
- Komarek J, Holy J (1999). Determination of heavy metals by electrothermal atomic absorption spectrometry after electrodeposition on a graphite probe. Spectrochim Acta B-Atomic Spectro 54: 733-738.
- Nutiu R, Li Y (2005). In vitro selection of structure-switching signaling aptamers. Angew Chem-Int Ed Engl 44: 1061-1065.
- Li F, Zhang H, Wang Z, Newbigging AM, Reid MS, et al. (2015) Aptamers facilitating amplified detection of biomolecules. Anal Chem 87: 274-292.
- Nutiu R, Li Y (2003) Structure-switching signaling aptamers. J Am Chem Soc 125: 4771-4778.
- Jhaveri SD, Kirby R, Conrad R, Maglott EJ, Bowser M, et al. (2000) Designed signaling aptamers that transduce molecular recognition to changes in fluorescence intensity. J Am Chem Soc 122: 2469-2473.
- Zhao Q, Geng X, Wang H (2013) Fluorescent sensing ochratoxin A with single fluorophore-labeled aptamer. Anal Bioanal Chem 405: 6281-6286.
- Cho E, Lee JW, Ellington AD (2009) Applications of aptamers as sensors. Annu Rev Anal Chem (Polo Alto Calif) 2: 241-264.
- Deng B, Lin Y, Wang C, Li F, Wang Z, et al. (2014) Aptamer binding assays for proteins: the thrombin example-a review. Anal Chim Acta 837: 1-15.
- Zheng J, Yang R, Shi M, Wu C, Fang X, et al. (2015) Rationally designed molecular beacons for bioanalytical and biomedical applications. Chem Soc Rev 44: 3036-3055.
- Sanzani SM, Reverberi M, Fanelli C, Ippolito A (2015) Detection of ochratoxin A using molecular beacons and real-time PCR thermal cycler. Toxins (Basel) 7: 812-820.
- Huang K, Marti AA (2012) Recent trends in molecular beacon design and applications. Anal Bioanal Chem 402: 3091-3102.
- Nutiu R, Li Y (2004) Structure-switching signaling aptamers: transducing molecular recognition into fluorescence signaling. Chemistry 10: 1868-1876.
- Chen J, Fang Z, Liu J, Zeng L (2012) A simple and rapid biosensor for ochratoxin A based on a structure-switching signaling aptamer. Food Cont 25: 555-560.
- Sharma A, Catanante G, Hayat A, Istamboulie G, Rejeb BI, et al. (2016) Development of structure switching aptamer assay for detection of aflatoxin M1 in milk samples. Talanta 158: 35-41.
- Dziuba D, Pohl R, Hocek M (2014) Bodipy-labeled nucleoside triphosphates for polymerase synthesis of fluorescent DNA. Bioconjug Chem 25: 1984-1995.
- Tanpure AA, Pawar MG, Srivatsan SG (2013) Fluorescent nucleoside analogs: probes for investigating nucleic acid structure and function. Israel J Chem 53: 366-378.
- Wilson JN, Cho Y, Tan S, Cuppoletti A, Kool ET (2008) Quenching of fluorescent nucleobases by neighboring DNA: the "insulator" concept. Chembiochem 9: 279-285.
- Manderville RA, Omumi A, Rankin KM, Wilson KA, Millen AL, et al. (2012) Fluorescent C-linked C8-aryl-guanine probe for distinguishing syn from anti structures in duplex DNA. Chem Res Toxicol 25: 1271-1282.
- Wilson JN, Kool ET (2006) Fluorescent DNA base replacements: reporters and sensors for biological systems. Org Biomol Chem 4: 4265-4274.
- Sproviero M, Manderville RA (2014) Harnessing G-tetrad scaffolds within G-quadruplex forming aptamers for fluorescence detection strategies. Chem Comm 50: 3097-3099.
- Jones AC, Neely RK (2015) 2-Aminopurine as a fluorescent probe of DNA conformation and the DNA-enzyme interface. Q Rev Biophys 48: 244-279.
- Vummidi BR, Alzeer J, Luedtke NW (2013) Fluorescent probes for G-quadruplex structures. Chembiochem 14: 540-558.
- Blanchard DJ, Cservenyi TZ, Manderville RA (2015) Dual fluorescent deoxyguanosine mimics for FRET detection of G-quadruplex folding. Chem Commun (Camb) 51: 16829-16831.
- Blanchard DJM, Fadock KL, Sproviero M, Deore PS, Cservenyi TZ, et al. (2016) Photophysical properties of push-pull 8-aryl-deoxyguanosine probes within duplex and G-quadruplex structures. J Mat Chem C 4: 2915-2924.
- Fadock KL, Manderville RA, Sharma P, Wetmore SD (2016) Optimization of fluorescent 8-heteroaryl-guanine probes for monitoring protein-mediated duplex-G-quadruplex exchange. Org Biomol Chem 14: 4409-4419.
- Park H, Paeng IR (2011) Development of direct competitive enzyme-linked aptamer assay for determination of dopamine in serum. Anal Chim Acta 685: 65-73.
- Fu P, Sun Z, Yu Z, Zhang Y, Shen J, et al. (2014) Enzyme linked aptamer assay: based on a competition format for sensitive detection of antibodies to mycoplasma bovis in serum. Anal Chem 86: 1701-1709.
- Park JH, Cho YS, Kang S, Lee EJ, Lee GH, et al. (2014) A colorimetric sandwich-type assay for sensitive thrombin detection based on enzyme-linked aptamer assay. Anal Biochem 462: 10-12.
- Toh SY, Citartan M, Gopinath SC, Tang TH (2015) Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens Bioelectron 64: 392-403.
- Barthelmebs L, Jonca J, Hayat A, Prieto-Simon B, Jean-Louis M (2011) Enzyme-linked aptamer assays (ELAAs), based on a competition format for a rapid and sensitive detection of ochratoxin A in wine. Food Cont 22: 737-743.
- Bonel L, Vidal JC, Duato P, Castillo JR (2011) An electrochemical competitive biosensor for ochratoxin A based on a DNA biotinylated aptamer. Biosens Bioelectron 26: 3254-3259.
- Gu C, Long F, Zhou X, Shi H (2016) Portable detection of ochratoxin A in red wine based on a structure-switching aptamer using a personal glucometer. RSC Adv 6: 29563-29569.
- Willner I, Shlyahovsky B, Zayats M, Willner B (2008) DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem Soc Rev 37: 1153-1165.
- Yang DK, Kuo CJ, Chen LC (2015) Synthetic multivalent DNAzymes for enhanced hydrogen peroxide catalysis and sensitive colorimetric glucose detection. Anal Chim Acta 856: 96-102.
- Shim WB, Mun H, Joung HA, Ofori JA, Chung DH, et al. (2014) Chemiluminescence competitive aptamer assay for the detection of aflatoxin B1 in corn samples. Food Cont 36: 30-35.
- Yang C, Lates V, Prieto-Simon B, Marty JL, Yang X (2013) Rapid high-throughput analysis of ochratoxin A by the self-assembly of DNAzyme-aptamer conjugates in wine. Talanta 116: 520-526.
- Lee J, Jeon CH, Ahn SJ, Ha TH (2014) Highly stable colorimetric aptamer sensors for detection of ochratoxin A through optimizing the sequence with the covalent conjugation of hemin. Analyst 139: 1622-1627.
- Seok Y, Byun JY, Shim WB, Kim MG (2015) A structure-switchable aptasensor for aflatoxin B1 detection based on assembly of an aptamer/split DNAzyme. Anal Chim Acta 886: 182-187.
- Freeman R, Liu X, Willner I (2011) Chemiluminescent and chemiluminescence resonance energy transfer (CRET) detection of DNA, metal ions, and aptamer-substrate complexes using hemin/G-quadruplexes and CdSe/ZnS quantum dots. J Am Chem Soc 133: 11597-11604.
- Luo M, Chen X, Zhou G, Xiang X, Chen L, et al. (2012) Chemiluminescence biosensors for DNA detection using graphene oxide and a horseradish peroxidase-mimicking DNAzyme. Chem Commun (Camb) 48: 1126-1128.
- Jo EJ, Mun H, Kim SJ, Shim WB, Kim MG (2016) Detection of ochratoxin A (OTA) in coffee using chemiluminescence resonance energy transfer (CRET) aptasensor. Food Chem 194: 1102-1107.
- Yang L, Jin M, Du P, Chen G, Zhang C, et al. (2015) Study on enhancement principle and stabilization for the luminol-H2O2-HRP chemiluminescence system. PloS One 10: e0131193.
- Wang C, Dong X, Liu Q, Wang K (2015) Label-free colorimetric aptasensor for sensitive detection of ochratoxin A utilizing hybridization chain reaction. Anal Chim Acta 860: 83-88.
- Heerema SJ, Dekker C (2016) Graphene nanodevices for DNA sequencing. Nature Nanotech 11: 127-136.
- Wu S, Duan N, Zhang W, Zhao S, Wang Z (2016) Screening and development of DNA aptamers as capture probes for colorimetric detection of patulin. Anal Biochem 508: 58-64.
- Lu Z, Chen X, Wang Y, Zheng X, Li CM (2015) Aptamer based fluorescence recovery assay for aflatoxin B1 using a quencher system composed of quantum dots and graphene oxide. Microchim Acta 182: 571-578.
- Lv L, Cui C, Liang C, Quan W, Wang S, et al. (2016) Aptamer-based single-walled carbon nanohorn sensors for ochratoxin A detection. Food Cont 60: 296-301.
- Liu B, Huang PJ, Kelly EY, J L (2016) Graphene oxide surface blocking agents can increase the DNA biosensor sensitivity. Biotechnol J 11: 780-787.
- Zhang J, Li Z, Zhao S, Lu Y (2016) Size-dependent modulation of graphene oxide-aptamer interactions for an amplified fluorescence-based detection of aflatoxin B1 with a tunable dynamic range. Analyst 141: 4029-4034.
- Zhang J, Zhang X, Yang G, Chen J, Wang S (2013) A signal-on fluorescent aptasensor based on Tb3+ and structure-switching aptamer for label-free detection of ochratoxin A in wheat. Biosens Bioelectron 41: 704-709.
- Costantini F, Sberna C, Petrucci G, Reverberi M, Domenici F, et al. (2016) Aptamer-based sandwich assay for on chip detection of ochratoxin A by an array of amorphous silicon photosensors. Sens Actuat B: Chem 230: 31-39.
- Liu R, Huang Y, Ma Y, Jia S, Gao M, et al. (2015) Design and synthesis of target-responsive aptamer-cross-linked hydrogel for visual quantitative detection of ochratoxin A. ACS Appl Mat Interfaces 7: 6982-6990.
- Castillo G, Spinella K, Poturnayova A, Snejdarkova M, Mosiello L, et al. (2015) Detection of aflatoxin B1 by aptamer-based biosensor using PAMAM dendrimers as immobilization platform. Food Cont 52: 9-18.
- Zhao Y, Yang Y, Luo Y, Yang X, Li M, Song Q (2015) Double detection of mycotoxins based on SERS labels embedded Ag@Au core-shell nanoparticles. ACS Appl Mater Interfaces 7: 21780-21786.
- Zhang J, Yao-Kun X, Chen M, Dong-Zhi W, Shu-Xian C, et al. (2016) A fluorescent aptasensor based on DNA-scaffolded silver nanoclusters coupling with Zn (II)-ion signal-enhancement for simultaneous detection of OTA and AFB1. Sens Actuat B: Chem 235: 79-85.
- Chen X, Bai X, Li H, Zhang B (2015) Aptamer-based microcantilever array biosensor for detection of fumonisin B-1. RSC Adv 5: 35448-35452.
- Soh JH, Lin Y, Rana S, Ying JY, Stevens MM (2015) Colorimetric detection of small molecules in complex matrixes via target-mediated growth of aptamer-functionalized gold nanoparticles. Anal Chem 87: 7644-7652.
- Zhu Z, Peng M, Zuo L, Zhu Z, Wang F, et al. (2015) An aptamer based surface plasmon resonance biosensor for the detection of ochratoxin A in wine and peanut oil. Biosens Bioelectron 65: 320-326.
- Huang KJ, Shuai HL, Chen YX (2016) Layered molybdenum selenide stacking flower-like nanostructure coupled with guanine-rich DNA sequence for ultrasensitive ochratoxin A aptasensor application. Sens Actuat B: Chem 225: 391-397.
- Lu L, Wang M, Liu LJ, Leung CH, Ma DL (2015) Label-free luminescent switch-on probe for ochratoxin A detection using a G-quadruplex-selective iridium(III) complex. ACS Appl Mater Interfaces 7: 8313-8318.
- Spinella K, Mosiello L, Poturnayova A, Sneyedarkova M, Hianik T, et al. (2015) Acoustic aptasensor or aflatoxin B1 determination. Sensors 319: 71-75.
- Jianga HL, Liua XY, Qiua YX, Yaob DS, Xiea CF, et al. (2015) Development of an aptasensor for the fast detection of Versicolorin A. Food Cont 56: 202-210.
- Zhou W, Kong W, Dou X, Zhao M, Ouyang Z, et al. (2016) An aptamer based lateral flow strip for on-site rapid detection of ochratoxin A in Astragalus membranaceus. J Chromatogr B Analyt Technol Biomed Life Sci 1022: 102-108.
- Ono A, Togashi H (2004) Highly selective oligonucleotide-based sensor for mercury(II) in aqueous solutions. Angew Chem-Int Ed Engl 43: 4300-4302.
- Dairaku T, Furuita K, Sato H, Sebera J, Yamanaka D, et al. (2016) Direct detection of the mercury-nitrogen bond in the thymine-Hg(II)-thymine base-pair with (199)Hg NMR spectroscopy. Chem Commun (Camb) 51: 8488-8491.
- Lippert B, Sanz Miguel PJ (2016) The renaissance of metal-pyrimidine nucleobase coordination chemistry. Acc Chem Res 49: 1537-1545.
- Aragay G, Pons J, Merkoci A (2011) Recent trends in macro-, micro-, and nanomaterial-based tools and strategies for heavy-metal detection. Chem Rev 111: 3433-3458.
- Wang R, Zhou X, Shi H, Luo Y (2016) T-T mismatch-driven biosensor using triple functional DNA-protein conjugates for facile detection of Hg2+. Biosens Bioelectron 78: 418-422.
- Li J, Sun M, Wei X, Zhang L, Zhang Y (2015) An electrochemical aptamer biosensor based on "gate-controlled" effect using β-cyclodextrin for ultra-sensitive detection of trace mercury. Biosens Bioelectron 74: 423-426.
- Wang W, Kang TS, Chan PW, Lu JJ, Chen XP, et al. (2015) A label-free G-quadruplex-based mercury detection assay employing the exonuclease III-mediated cleavage of T-Hg2+-T mismatched DNA. Sci Technol Adv Mater 16: 065004.
- Ruscito A, DeRosa MC (2016) Small-molecule binding aptamers: selection strategies, characterization, and applications. Front Chem 4: 14.
- Wang H, Cheng H, Wang J, Xu L, Chen H, et al. (2016) Selection and characterization of DNA aptamers for the development of light-up biosensor to detect Cd(II). Talanta 154: 498-503.
- Qian ZS, Shan XY, Chai LJ, Chen JR, Feng H (2015) A fluorescent nanosensor based on graphene quantum dots-aptamer probe and graphene oxide platform for detection of lead (II) ion. Biosens Bioelectron 68: 225-231.
- Cui L, Wu J, Ju H (2016) Label-free signal-on aptasensor for sensitive electrochemical detection of arsenite. Biosens Bioelectron 79: 861-865.
- Zhang L, Wong JX, Li X, Li Y, Yi HZ (2015) Detection and quantitation of heavy metal ions on bona fide DVDs using DNA molecular beacon probes. Anal Chem 87: 5062-5067.
- Tang X, Wang YS, Xue JH, Zhou B, Cao JX, et al. (2015) A novel strategy for dual-channel detection of metallothioneins and mercury based on the conformational switching of functional chimera aptamer. J Pharm Biomed Anal 107: 258-264.
- Liu W, Zhu H, Zheng B, Cheng S, Fu Y, et al. (2012) Kinetics and mechanism of G-quadruplex formation and conformational switch in a G-quadruplex of PS2.M induced by Pb2+. Nucleic Acids Res 40: 4229-4236.
- Wang H, Wang D, Huang CZ (2015) Highly sensitive chemiluminescent detection of lead ion based on its displacement of potassium in G-quadruplex DNAzyme. Analyst 140: 5742-5747.
- Liu T, Nie G, Zhang X, Liu W, Zhang W, Wang Y, et al. (2015) Development of a detection kit based on G-quadruplex DNAzyme for detection of lead(II) ion in food samples. Food Anal Methods 8: 1133-1140.
- Santoro SW, Joyce GF (1997) A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci U S A 94: 4262-4266.
- Zhang Y, Xiao S, Li H, Liu H, Pang P, et al. (2016) A Pb2+-ion electrochemical biosensor based on single-stranded DNAzyme catalytic beacon. Sens Actuat B: Chem 222: 1083-1089.
- Zhou Q, Lin Y, Lin Y, Wei Q, Chen G, et al. (2016) Highly sensitive electrochemical sensing platform for lead ion based on synergetic catalysis of DNAzyme and Au-Pd porous bimetallic nanostructures. Biosens Bioelectron 78: 236-243.
- Wang C, Cui X, Li Y, Li H, Huang L, et al. (2016) A label-free and portable graphene FET aptasensor for children blood lead detection. Sci Rep 6: 21711.
- Huang PJ, Liu J (2015) Rational evolution of Cd2+-specific DNAzymes with phosphorothioate modified cleavage junction and Cd2+ sensing. Nucleic Acids Res 43: 6125-6133.
- Tsekenis G, Filippidou MK, Chatzipetrou M, Tsouti V, Zergioti I, et al. (2015) Heavy metal ion detection using a capacitive micromechanical biosensor array for environmental monitoring. Sens Actuat B: Chem 208: 628-635.
- Zhang J, Tang Y, Teng L, Lu M, Tang D (2015) Low-cost and highly efficient DNA biosensor for heavy metal ion using specific DNAzyme-modified microplate and portable glucometer-based detection mOther designs using aptamers for mycotoxin detectionsode. Biosens Bioelectron 68: 232-238.
- Li C, Wei L, Liu X, Lei L, Li G (2014) Ultrasensitive detection of lead ion based on target induced assembly of DNAzyme modified gold nanoparticle and graphene oxide. Anal Chim Acta 831: 60-64.
- McKeague M, DeRosa MC (2012) Challenges and opportunities for small molecule aptamer development. J Nucleic Acids 2012: 748913.