Journal of Toxins
Download PDF
Research Article
*Address for Correspondence: Richard A Manderville, Departments of Chemistry and Toxicology, University of Guelph, Canada N1G 2W1, Tel: (519)-824-4120, x53963; Fax: (519)-766-1499; E-mail: rmanderv@uoguelph.ca
Citation: Verwey AMR, Witham AA, Li M, Manderville RA, et al. Mutagenicity Analysis of C8-Phenoxy-Guanine in the NarI Recognition DNA Sequence. J Toxins. 2014;1(1): 6.
Copyright © 2014 Manderville et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Toxins | ISSN: 2328-1723 | Volume: 1, Issue: 1
Submission: 31 July 2014 | Accepted: 06 August 2014 | Published: 11 August 2014
Reviewed & Approved by: Dr. Guangming Xiong, Head of the Pharmacology and Toxicology, University Kiel, Germany.
We recently incorporated C8-phenoxy-2′-deoxyguanine (PhOdG) into the G3 position of the NarI recognition sequence within a 12-mer oligonucleotide (5′-CTCGGCXCCATC, G3 = X = PhOdG) in order to study its structural characteristics to predict mutagenic outcome [23]. The NarI sequence is a hot-spot for frameshift mutations mediated by the N-linked C8-dG adduct of N-acetyl- 2-aminofluorene (AAF) [19-21]. Within the NarI(12) duplex, the PhOdG lesion was shown to adopt the major groove B conformation opposite C, with minimal disruption to the duplex structure [23]. These findings correlated with the conformational preference for the corresponding single-ringed N-linked C8-dG adduct produced by aniline [24]. On the basis of this comparison, the PhOdG lesion was predicted to be weakly mutagenic [23].
Translesion synthesis by Dpo4
In the presence of all four dNTPs (Figure 5), the unmodified template was fully extended to the 22-mer complementary strand by 5 min of incubation with Dpo4, with some additional incorporation of an 8th base for one base additional extension from a blunt end [29].
Mutagenicity Analysis of C8-Phenoxy-Guanine in the NarI Recognition DNA Sequence
Anne MR Verwey1, Aaron A Witham1, Mei Li2 and Richard A Manderville1*
- 1Departments of Chemistry and Toxicology, University of Guelph, Canada
- 2State Key Laboratory of Pollution Control and Resource Reuse, School of the Environment, Nanjing University, PR China
*Address for Correspondence: Richard A Manderville, Departments of Chemistry and Toxicology, University of Guelph, Canada N1G 2W1, Tel: (519)-824-4120, x53963; Fax: (519)-766-1499; E-mail: rmanderv@uoguelph.ca
Citation: Verwey AMR, Witham AA, Li M, Manderville RA, et al. Mutagenicity Analysis of C8-Phenoxy-Guanine in the NarI Recognition DNA Sequence. J Toxins. 2014;1(1): 6.
Copyright © 2014 Manderville et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Toxins | ISSN: 2328-1723 | Volume: 1, Issue: 1
Submission: 31 July 2014 | Accepted: 06 August 2014 | Published: 11 August 2014
Reviewed & Approved by: Dr. Guangming Xiong, Head of the Pharmacology and Toxicology, University Kiel, Germany.
Abstract
Phenoxyl radicals can covalently attach to the C8-site of 2′-deoxyguanosine (dG) to generate oxygen-linked biaryl ether C8-dG adducts. To determine the mutagenicity of an O-linked C8- dG adduct, C8-phenoxy-dG (PhOdG) was incorporated into the G3 position (X) of the NarI recognition sequence within a 22-mer oligonucleotide template (5′-CTCGGCX-CCATCCCTTACGAGC, where X = dG, or PhOdG) using solid-phase DNA synthesis. The NarI(22) template was annealed to a 15-mer primer and in vitro mutagenicity was assessed using primer-extension assays with a high-fidelity replicative polymerase, Escherichia coli pol I Klenow fragment exo− (Kf−), and a lesion-bypass Y-family polymerase, Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4). These studies predict that the O-linked C8-dG lesion PhOdG will have a low mutagenic effect, and is unlikely to contribute strongly to phenol toxicity.Keywords
Iron; Copper; Zinc; Manganese; Meat; Meat products; ICP-OESAbbreviations
dG: 2′-deoxyguanosine; PhOdG: C8-(phenoxy)-2′-deoxyguanosine; ROS: Reactive oxygen species; Kf−: Escherichia coli pol I Klenow fragment exo−; Dpo4: Y-family lesion-bypass DNA polymerase IVIntroduction
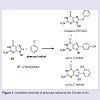
Phenols are ubiquitous compounds that possess many biological properties including toxicity [1-3]. Human exposure to phenolic toxins occurs predominately through industrial activities, tobacco smoke, and inhalation of polluted air [4,5]. Phenol toxicity stems from their oxidative metabolism by peroxidase or cytochrome P450 enzymes to generate hydroquinone/quinone redox pairs, reactive oxygen species (ROS) and phenoxyl radicals that are well-known DNA-damaging agents [1,6-8]. Human exposure to damaging radicals [9,10] and quinone electrophiles [6,7] are associated with cancer and aging.Our interest in phenol toxicity stems from the finding that certain phenolic radicals can attach covalently to the C8-site of 2′-deoxyguanosine (dG) to afford C8-dG adducts [11-14]. Due to the ambident electrophilicity of phenoxyl radicals (C vs O) both carbon-and oxygen-linked C8-dG adducts have been observed (Figure 1). The phenolic C-linked C8-dG adducts belong to a larger class of C8-aryl-dG lesions produced by a number of chemical mutagens that include polycyclic aromatic hydrocarbons (PAHs) [15], estrogens [16], nitroaromatics [17] and arylhydrazines [18]. The phenolic O-linked C8-dG adducts are unique to phenol precursors, but are expected to be structurally similar to the corresponding nitrogen-linked C8- dG adducts produced by aromatic amine carcinogens [19-21]. For aromatic amines, structure-activity relationships have demonstrated that all of the potent carcinogens are polycyclic structures, while none of the single-ringed aniline derivatives are potent carcinogens [22].
We recently incorporated C8-phenoxy-2′-deoxyguanine (PhOdG) into the G3 position of the NarI recognition sequence within a 12-mer oligonucleotide (5′-CTCGGCXCCATC, G3 = X = PhOdG) in order to study its structural characteristics to predict mutagenic outcome [23]. The NarI sequence is a hot-spot for frameshift mutations mediated by the N-linked C8-dG adduct of N-acetyl- 2-aminofluorene (AAF) [19-21]. Within the NarI(12) duplex, the PhOdG lesion was shown to adopt the major groove B conformation opposite C, with minimal disruption to the duplex structure [23]. These findings correlated with the conformational preference for the corresponding single-ringed N-linked C8-dG adduct produced by aniline [24]. On the basis of this comparison, the PhOdG lesion was predicted to be weakly mutagenic [23].
In the present study we utilize primer-extension assays to more accurately predict the mutagenic potential of the O-linked PhOdG adduct. In our model the PhOdG adduct has been incorporated into the G3 position of a NarI(22) template (5′-CTCGGCXCCAT-CCCTTACGAGC, where X = dG, or PhOdG) and annealed to a 15- mer primer strand (5′-GCTCGTAAGGGATGG). The bypass ability in vitro of the high-fidelity replicative DNA polymerase – E. coli DNA polymerase I Klenow fragment (exonuclease negative mutant, Kf−) was compared to bypass by the Y-family DNA polymerase IV (Dpo4) from Sulfolobus solfataricus [25]. High-fidelity polymerases utilize an ‘induced-fit’ mechanism of replication, and bulky adducts in the template strand often stall or block the progression of replication through distortion of the polymerase active site [26]. Such stalling in vivo is believed to be a trigger for the recruitment of Y-family translesion polymerases [26], which have a larger solvent-exposed active site to accommodate bulky DNA lesions for more probable effective bypass [27]. Mutagenesis experiments in vitro support our predictions derived from structural analysis of the PhOdG lesion within NarI(12) [23], and suggest that the O-linked C8-dG adduct is unlikely to contribute strongly to phenol toxicity.
Materials and Methods
MaterialsThe NarI(22) oligonucleotide substrates were prepared on a BioAutomation MerMade 12 automatic DNA synthesizer using standard or modified β-cyanoethylphosphoramidite chemistry (1 μmol scale). Full synthetic details of the PhOdG phosphoramidite have been previously published [23]. The 15-mer primer oligonucleotide was purchased from Sigma Genosys (Oakville, ON, Canada) and was purified by Sigma using polyacrylamide gel electrophoresis (PAGE). The purity of the oligonucleotide sample was checked at the University of Guelph using HPLC (see Supporting Information for HPLC analysis of the 15-mer primer (Figure S1)). Escherichia coli pol I Klenow fragment exo− (Kf−) and T4 polynucleotide kinase were purchased from New England BioLabs, while Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) was purchased from Trevigen Inc; isotopically-labeled ATP ([γ-32P]-ATP) was purchased from PerkinElmer.
Purification of Oligonucleotides
Aqueous solutions containing impure NarI(22) oligonucleotides were initially filtered using Mandel syringe filters (PVDF 0.20 μm). Oligonucleotide purification was carried out on an Agilent 1200 series HPLC equipped with an autosampler, diode array detector (monitored at λabs = 258 nm) and an autocollector. Separation was carried out at 50°C using a Phenomenex clarity 5 μ oligo-RP semi-preparative column (100 x 10 mm) at a flow rate of 3.5 mL/min using a gradient running from 95% 50 mM aqueous triethylamine acetate (TEAA, pH 7.2):5% acetonitrile to 30% 50 mM TEAA:70% acetonitrile over 30 minutes. The modified oligonucleotide sample was lyophilized to dryness and obtained as white foam.
DNA Quantification
Stock solutions of DNA (0.50 mL or 1.00 mL) were prepared in purified water. Consecutive additions of 5 μL of DNA stock to 1985 μL of purified water followed by an absorption scan at 260 nm on a Cary 300-Bio UV-Visible spectrophotometer equipped with a Peltier block-heating unit and automated temperature controller using standard 10 mm light path quartz glass cells from Hellma GmbH & Co (Concord, ON). Scans were performed three times in order to determine the concentration of the stock solution. Molar absorptivities (ε) of the unmodified NarI(22) were used for the modified NarI(22) strand and were calculated online using Integrated DNA Technologies (IDT) OligoAnalyzer 3.1.
Mass Spectrometry
MS experiments for DNA identification were conducted on a Bruker AmaZon quadrupole ion trap SL spectrometer. Masses were acquired in the negative ionization mode with an electrospray ionization source (Bruker Daltronics, Milton, Canada). The NarI(22) oligonucleotide sample was dissolved in 90% milliQ filtered water/10% MeOH with 0.1% ammonium acetate and injected directly into the electrospray source. Ionization analysis was carried out using the following settings on the ESI: nebulizer gas flow 40 psi, dry gas 10 L/min, dry temperature 200ºC, spray voltage was 4000 V. Scan range was 70-2000 m/z and scan resolution was 8100 m/z/s. Injection rate was 40μL/s. See Supporting Information for ESI-MS analysis (Table S1) and spectra (Figure S2) of the modified NarI(22) oligonucleotide.
Radiolabeling and Annealing
T4 polynucleotide kinase and [γ-32P] ATP were used to label the 15mer primer strands at the 5′-end. The unmodified and modified DNA template: primer duplexes were prepared by annealing the 15mer primer and NarI(22) template strands (50% excess of template strand) through heating the mixtures to 95ºC for ten minutes, followed by slow cooling to room temperature overnight.
Single nucleotide incorporation assays
Kf− or Dpo4 were used to perform primer extension reactions on each previously labeled and annealed template: Primer duplex in the presence of 25 μM of dCTP, dGTP, dATP, or dTTP. Reactions were initiated by the addition of the dNTP to enzyme/DNA mixtures to give a final reaction volume of 10 μL. The final concentrations for Kf− assays were 50 mM NaCl, 10mM Tris-HCl (pH 7.9), 10mM MgCl2, 1mM dithiothreitol (DTT), 100 nM duplex, and 20 nM Kf−. The final concentrations for Dpo4 assays were 50 mM NaCl, 50 mM Tris (pH 8.0), 2.5 mM MgCl2, 5 mM DTT, 100 μg/mL bovine serum albumin (BSA), 5% glycerol, 100 nM duplex, and 20 nM Dpo4. Reactions were incubated at 37ºC for 1 hour with Kf−, and 30 minutes with Dpo4, followed by 4 μL transferred and mixed with 36 μL of loading dye (95% formamide, 20 mM EDTA, 0.05% xylene cyanol and bromophenol blue) to terminate the reaction. 4 μL of these quenched reactions were then subjected to 15% polyacrylamide gel electrophoresis in the presence of 7 M urea and incorporation products were visualized using a Bio-Rad phosphorimager. Relative frequency of base incorporation was determined using ImageJ64 software.
Full length extension assays
Kf− or Dpo4 were used to perform primer extension reactions on each previously labeled and annealed template:primer duplex in the presence of a 100 μM 4dNTP mix (25 μM each dNTP). A final concentration of 20 nM of Kf− or Dpo4 was used in each reaction, while all other final concentrations were the same as outlined for the single nucleotide incorporation assays. Reactions were initiated by the addition of the 4dNTP mix to enzyme/DNA mixtures to give a final reaction volume of 10 μL with Kf−, or 20 μL with Dpo4. Reactions with Kf− were incubated at 37ºC for 1 hour, and then quenched in the same manner as described above. Reactions with Dpo4 were also incubated at 37ºC, but 4 μL aliquots were removed from incubation after 2, 5, 10, 15, and 30 minutes with the unmodified duplex, and after 15, 30, 45, 60, and 90 minutes with the modified duplex. Each 4 μL aliquot removed was quenched in the same manner as listed above. All quenched reaction samples were then subjected to 15% polyacrylamide gel electrophoresis in the presence of 7 M urea and incorporation products were visualized using a Bio-Rad phosphorimager.
Results and Discussion
Primer extension by Klenow fragment exo− (Kf−) Single nucleotide insertion assays using Kf− were carried out using the NarI(22):15mer template:primer (Figure 2). On the unmodified NarI(22) template (X = dG), Kf− mainly incorporated the correct base C (~ 82%, Figure 2b). At 20 nM enzyme concentration, significant amounts of G (~37%) and T (~30%) were also incorporated opposite dG during the 60 min reaction time. On the modified NarI(22) template (X = PhOdG), the polymerase again mainly inserted the correct base C (~70%, Figure 2b). However, a higher relative frequency of a second C (10%) and a slight increase in the insertion of A compared to the unmodified template was observed. The PhOdG adduct also inhibited misincorporation of G and T compared to the unmodified template (Figure 2b).Figure 2: a) Single nucleotide incorporation primer extension assays with 25μM of individual dNTPs as indicated under each lane, by incubation of 20 nM of Kf− with each duplex (sequence presented above gel) for 1 hour at 37°C. (Note: B = blank where no enzyme present). b) Relative frequency of each dNTP incorporated by Kf− on NarI(22):15-mer template:primer duplex where X=dG (black), or X=PhOdG (grey).
In the presence of all four dNTPs (Figure 3), extension past PhOdG was stalled after the incorporation of one nucleotide (presumably C across from PhOdG), though the full length extension product was clearly obtained. The observed stalling at position 1 (numbering on gel in Figure 3) is in agreement with previous reports that extension past bulky C8-dG adducts is more difficult than base insertion opposite the lesion [28]. An 8th band for one base additional extension beyond the template strand was also observed, which is a typical non-template-dependent extension from a blunt end [29].
Figure 3: a) Full length primer extension assays on NarI(22):15-mer template:primer duplexes (sequence presented above gel) where X=dG or PhOdG by 20 nM Kf− in the presence of 25 μM of each dNTP. Reactions were incubated for 60 min at 37°C. The blank sample had no enzyme present during the reaction with unmodified duplex.
Given that the PhOdG adduct stalls replication by the high-fidelity polymerase Kf−, it was desirable to examine extension using a lesion-bypass DNA polymerase. Distortions and attendant stalling of high-fidelity polymerases by bulky adducts are regarded as signals for recruiting lesion-bypass polymerases in vivo [26]. Thus, bulky phenolic adducts are more likely to be processed by translesion polymerases and to model this interaction we employed the Y-family polymerase, Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) [25,26].
Single nucleotide insertion assays using Dpo4 were carried out using the NarI(22):15mer template:primer (Figure 4). On both the unmodified and modified NarI(22) templated, Dpo4 exhibits low-fidelity with significant amounts of misincorporations (Figure 4b). For unmodified templates, this phenomenon is known for translesion polymerases due to decreased interactions of the larger, more spacious active site with the template DNA, providing lower geometric selection for correct base pairs [30]. Typically, the Y-family polymerases will exhibit much higher fidelity when replicating damaged DNA [31]. However, in this instance the PhOdG adduct had little impact on the fidelity of Dpo4 compared to the unmodified control (Figure 4b).
Figure 4: a) a) Single nucleotide incorporation primer extension assays with 25μM of individual dNTPs as indicated under each lane, by incubation of 20 nM of Dpo4 with each duplex (sequence presented above gel) for 30 min at 37°C. (Note: B = blank where no enzyme present). b) Relative frequency of each dNTP incorporated by Dpo4 on NarI(22):15-mer template:primer duplex where X=dG (black), or X=PhOdG (grey).
Figure 5: a) Full length primer extension assays on NarI(22):15-mer template: primer duplexes (sequence presented above gel) where X=dG or PhOdG by 20 nM Dpo4 in the presence of 25 μM of each dNTP. Aliquotsof each reaction were removed from incubation at 37°C after the time listed under each lane. (Note: B = blank where no enzyme present).
Conclusions
The current study has allowed us to conclude the following: (1) the single-ringed oxygen-linked C8-phenoxy-dG adduct (PhOdG) does not strongly impede the progress of DNA replication by either Kf− or Dpo4 when inserted into the NarI(22) template and annealed to a 15-mer primer. Both the high-fidelity polymerase Kf− and the lesion-bypass polymerase Dpo4 are able to fully extend the 15-mer primer in the presence of the PhOdG lesion, although PhOdG causes some stalling after the first incorporation opposite the adduct. (2) In single nucleotide insertion assays, the PhOdG adduct does not strongly alter the relative frequency of dNTP incorporation compared to insertion opposite dG. Overall, these results suggest that PhOdG is a weakly mutagenic lesion, which correlates with our earlier prediction for PhOdG based on its structural characteristics within the NarI(12) duplex. These findings suggest that the O-linked PhOdG adduct will not strongly contribute to phenol toxicity. Our results are the first to report the in vitro mutagenicity of an oxygen-linked biaryl ether C8-dG adduct and provide a basis for comparison to other O-linked C8-dG adducts derived from phenolic toxins.Supporting Information Available
HPLC chromatogram of 15-mer primer, ESI-MS analysis and spectra of the modified NarI oligonucleotide.
Acknowledgements
Support for this research was provided by the Natural Sciences and Engineering Research Council (NSERC) of Canada, the Canada Foundation for Innovation, the Ontario Innovation Trust Fund.References
- Manderville RA (2009) DNA damage by phenoxyl radicals: Radical and Radical Ion Reactivity in Nucleic Acid Chemistry (Greenberg M, Ed.) John Wiley & Sons, Hoboken, NJ 421-443.
- Shadnia H, Wright JS (2008) Understanding the toxicity of phenols: using quantitative structure-activity relationships and enthalpy changes to discriminate between possible mechanisms. Chem Res Toxicol 21: 1197-1204.
- Wright JS, Shadnia H (2008) Computational modeling of substituent effects on phenol toxicity. Chem Res Toxicol 21: 1426-1431.
- Weber M, Weber M, Kleine-Boymann M (2004) Phenol: Ullmann's Encyclopedia of Industrial Chemistry. John Wiley & Sons, New York.
- Smith CJ, Perfetti TA, Morton MJ, Rodgman A, Garg R, et al. (2002) The relative toxicity of substituted phenols reported in cigarette mainstream smoke. Toxicol Sci 69: 265-278.
- Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ (2000) Role of quinones in toxicology. Chem Res Toxicol 13: 135-160.
- Bolton JL (2002) Quinoids, quinoid radicals, and phenoxyl radicals formed from estrogens and antiestrogens. Toxicology 177: 55-65.
- Manderville RA, Pfohl-Leszkowicz A (2006) Genotoxicity of chlorophenols and ochratoxin A: Advances in Molecular Toxicology (Fishbein JC, Ed.) Elsevier, Amsterdam 85-138.
- Pryor WA (1997) Cirgarette-smoke radicals and the role of free radicals in chemical carcinogenicity. Environ Health Perspect 105: 875-882.
- Sandstead H (1995)Scarfiotti C, Fabris F, Cestaro B, Giuliani A (1997) Free radicals, clinical atherosclerosis, ageing, and related dysmetabolic pathologies: Pathological and aspects. Eur J Cancer Prev 6: S31-36.
- Dai J, Wright MW, Manderville RA (2003) Ochratoxin A forms a carbon-bonded C8-deoxyguanosine nucleoside adduct: Implication for C8-reactivity by a phenolic radical. J Am Chem Soc 125: 3716-3717.
- Dai J, Wright MW, Manderville RA (2003) An oxygen-bonded C8-deoxyguanosine nucleoside adduct of pentachlorophenol by peroxidase activation: evidence for ambident C8 reactivity by phenoxyl radicals. Chem Res Toxicol 16: 817-821.
- Dai J, Sloat AL, Wright MW, Manderville RA (2005) Role of phenoxyl radicals in DNA adduction by chlorophenol xenobiotics following peroxidase activation. Chem Res Toxicol 18: 771-779.
- Manderville RA (2005) Ambident reactivity of phenoxyl radicals in DNA adduction. Can J Chem 83: 1261-1267.
- Rogan EG, Cavalieri EL, Tibbels SR, Cremonesi P, Warner CD, et al. (1988) Synthesis and identification of benzo[a]pyrene-guanine nucleoside adducts formed by electrochemical oxidation and by horseradish peroxidase catalyzed reaction of benzo[a]pyrene with DNA. J Am Chem Soc 110: 4023-4029.
- Akanni A, Abul-Hajj YJ (1999) Estrogen-nucleic acid adducts: Dissection of the reaction of 3,4-estrone quinone and its radical anion and radical cation with deoxynucleosides and DNA. Chem Res Toxicol 12: 1247-1253.
- Enya T, Kawanishi M, Suzuki H, Matsui S, Hisamatsu Y (1998) An unusual DNA adduct derived from the powerfully mutagenic environmental contaminant 3-nitrobenzanthrone. Chem Res Toxicol 11: 1460-1467.
- Hiramoto K, Kaku M, Sueyoshi A, Fujise M, Kikugawa K (1995) DNA base and deoxyribose modification by the carbon-centered radical generated from 4-(hydroxymethyl) benzenediazonium salt, a carcinogen in mushroom. Chem Res Toxicol 8: 356-362.
- Hoffmann GR, and Fuchs RPP (1997) Mechanisms of frameshift mutations: Insight from aromatic amines. Chem Res Toxicol 10: 347-359.
- Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, et al. (1998) Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem Res Toxicol 11: 391-407.
- Cho B (2010) Structure-function characteristics of aromatic amine-DNA adducts: The Chemical Biology of DNA Damage 217-238.
- Josephy PD, Mannervik B (2006) Molecular Toxicology (2ndedn), Oxford University Press, New York, pp 517-518.
- Kuska MS, Witham AA, Sproviero M, Manderville RA, Majdi Yazdi M, et al. (2013) Structural influence of C8-phenoxy-guanine in the NarI recognition DNA sequence. Chem Res Toxicol 26: 1397-1408.
- Shapiro R, Ellis S, Hingerty BE, Broyde S (1998) Effect of ring size on conformations of aromatic amine–DNA adducts: The aniline–C8 guanine adduct resides in the B-DNA major groove. Chem Res Toxicol 11: 335-341.
- Guengerich FP (2006) Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem Rev 106: 420-452.
- Broyde S, Wang L, Rechkoblit O, Geacintov NE, Patel DJ (2008) Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem Sci 33: 209-219.
- Eoff RL, Choi JY, Guengerich FP (2010) Mechanistic studies with DNA polymerases reveal complex outcomes following bypass of DNA damage. J Nucleic Acids: ID 830473.
- Miller H, Grollman AP (1997) Kinetics of DNA polymerase I (Klenow fragment exo−) activity on damaged DNA templates: Effect of proximal and distal template damage on DNA synthesis. Biochemistry 36: 15336-15342.
- Kirouac KN, Basu AK, Ling H (2013) Structural mechanism of replication stalling on a bulky amino-polycyclic aromatic hydrocarbon DNA adduct by a Y family DNA polymerase. J Mol Biol 425: 4167-4176.
- Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA (2002) Low fidelity DNA synthesis by a Y family DNA polymerase due to misalignment in the active site. J Biol Chem 277: 19633-19638.
- Berdis AJ (2009) Mechanisms of DNA polymerases. Chem Rev 109: 2862-2879.