Journal of Orthopedics & Rheumatology
Download PDF
Research Article
*Address for Correspondence: Guangju Zhai, Discipline of Genetics, Faculty of Medicine, Memorial University of Newfoundland, St John’s, NL, Canada, A1B 3V6, Tel: 1 709 777 7286; Fax: 1 709 777 7497; Email: guangju.zhai@med.mun.ca
Citation: Aref-Eshghi E, Rahman P, Zhang H, Martin G, Furey A, et al. Attempt to replicate the published osteoarthritis-associated genetic variants in the Newfoundland & Labrador Population. J Orthopedics Rheumatol. 2014;1(3): 5.
Copyright © 2014 Aref-Eshghi E, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Orthopedics & Rheumatology | ISSN: 2334-2846 | Volume: 1, Issue: 3Submission: 14 April 2014 | Accepted: 17 April 2014 | Published: 23 April 2014
Reviewed & Approved by: Dr. Andrew P. Andonopoulos, Department of Internal Medicine and Rheumatology, Patras University School of Medicine, Greece
Attempt to replicate the published osteoarthritis associated genetic variants in the Newfoundland & Labrador Population
Erfan Aref-Eshghi1, Proton Rahman2, Hongwei Zhang1, Glynn Martin3, Andrew Furey3, RogerGreen1, Guang Sun2 and Guangju Zhai1,4*
- 1Discipline of Genetics, Faculty of Medicine, Memorial University of Newfoundland, Canada
- 2Discipline of Medicine, Faculty of Medicine, Memorial University of Newfoundland, Canada
- 3Division of Orthopaedics, Faculty of Medicine, Memorial University of Newfoundland, Canada
- 4Department of Twin Research & Genetic Epidemiology, King’s College London, UK
*Address for Correspondence: Guangju Zhai, Discipline of Genetics, Faculty of Medicine, Memorial University of Newfoundland, St John’s, NL, Canada, A1B 3V6, Tel: 1 709 777 7286; Fax: 1 709 777 7497; Email: guangju.zhai@med.mun.ca
Citation: Aref-Eshghi E, Rahman P, Zhang H, Martin G, Furey A, et al. Attempt to replicate the published osteoarthritis-associated genetic variants in the Newfoundland & Labrador Population. J Orthopedics Rheumatol. 2014;1(3): 5.
Copyright © 2014 Aref-Eshghi E, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Orthopedics & Rheumatology | ISSN: 2334-2846 | Volume: 1, Issue: 3Submission: 14 April 2014 | Accepted: 17 April 2014 | Published: 23 April 2014
Reviewed & Approved by: Dr. Andrew P. Andonopoulos, Department of Internal Medicine and Rheumatology, Patras University School of Medicine, Greece
Abstract
Objective: Over 200 genes have been reported to be associated with osteoarthritis (OA), but most of them have not been replicated in an independent sample. Using the newly collected cohort from a genetically isolated population - the Newfoundland and Labrador population, we attempted to replicate 105 previously reported OA-associated SNPs.Methods: A case-control study design was utilized in this study. Patients undergoing total hip/knee joint replacements due to severe OA were collected as cases. A group of healthy individuals with no evidence of OA was used as control. 105 SNPs were genotyped either by Sequenom iPLEX Gold method or Illumina GWAS genotyping platform. The cross-reference was performed on both methods in a subset of samples for genotyping quality control. A logistic regression model was used to test for associations between the SNPs and OA.
Results: A total of 126 cases and 348 healthy controls were included in the final analysis. OA Patients were on average 9 years older than healthy controls (p<0.0001), but there was no difference in BMI. We were unable to replicate the previously reported associations. Two SNPs, rs2294995 (COL9A3), and rs1049007 (BMP2) showed an association with p<0.05, but the significance did not survive the Bonferroni multiple testing correction.
Conclusion: A lack of replication might be due to study design, complexity of OA, method of OA ascertainment, populations studied, or false positives in the original publications. A study with larger sample is needed to confirm the two possible SNPs with OA.
Keywords
Osteoarthritis; Genetics; Single nucleotide polymorphism; ReplicationIntroduction
Osteoarthritis (OA) is the most common form of arthritis causing joint pain, stiffness, limited range of motion, joint deformity, and disability [1]. Its prevalence is on the rise due to population aging and increasing prevalence in obesity. Knee, hip, hand, spine, and foot are the most affected joints while the greatest public health burden results from hip and/or knee OA [2]. To date, there are no drugs available for rebuilding the damaged cartilage, nor is there a clear understanding of the pathogenesis of the disease. Total joint replacement therapy is the only choice for people with advanced OA. In the US alone, the total number of hip and/or knee joint replacement surgery due to OA is 350,000 each year [3], and the annual per person cost of those living with OA has been estimated to be around $5700 [4]. Arthritis, mostly OA, costs $128 million per year in medical care and other indirect expenses including those resulting from work limitation and loss of productivity in the US [5].Although the etiology of OA is not completely understood, it is believed that OA is a multifactorial condition developing and progressing as a result of a combination of different environmental and genetic factors [6]. The main environmental risk factors include age, gender, obesity, previous joint injury, and joint mal-alignments [7]. Evidence suggests a strong genetic component to OA; from twin studies this genetic influence has been estimated to be between 40% and 65% for knee, hip [8], and hand OA [9], and first-degree relatives of individuals with spine, hand, hip, or polyarticular OA have a OA have a two- to three-fold increased risk of the disease [10,11].
To date, 9 Genome-Wide Association Studies (GWAS), along with a large number of candidate gene studies and linkage analyses, have been performed on OA. Although some OA-associated genes such as GDF5 have been replicated in independent studies, the majority of the studied loci have inconsistent results [12]. This might be due to genetic heterogeneous nature of the disease or false positive findings in the initial studies. Genetically isolated populations have advantages for complex disease gene mapping because of their reduced genetic heterogeneity and extended LD [13]. The Newfoundland and Labrador population is a young isolated founder population with a high degree of both genetic and cultural homogeneity exhibiting extended linkage disequilibrium and an increased kinship coefficient, which provides a unique source for investigating both single gene diseases and complex traits [14,15]. Using this unique population, the aim of the present study was to replicate previously reported OA-associated genes.
Methods and Materials
SubjectsThe study was part of the Newfoundland Osteoarthritis Study (NFOAS) that was initiated in 2011 and aimed at identifying novel genetic, epigenetic, and biochemical markers for OA. OA patients were recruited from those who underwent total knee or hip replacement surgery due to primary OA between Nov. 2011 to Jun. 2013 in St. Clare’s Mercy Hospital and Health Science Centre General Hospital in St. John’s, the capital city of Newfoundland and Labrador (NL) province of Canada. A group of healthy people who do not have any evidence of either knee or hip OA were used as controls. The controls were selected from previous genetic association study for type 2 diabetes and obesity. The controls completed a questionnaire regarding any ongoing symptoms, previous diagnosis and medication history. Each patient was also examined by a rheumatologist. The controls were selected for this study if they had no musculoskeletal pain, a prior diagnosis of osteoarthritis, were not taking acetaminophen or NSAIDs and had a normal physical examination, as it relates to the skeletal system. All cases and controls in this study were from NL. The study was approved by Health Research Ethics Authority (HREA) of Newfoundland and Labrador and a written consent was obtained from all the participants.
Height and weight measurements were obtained from patient’s hospital medical record. Body mass index (BMI) was calculated as weight in kilogram divided by squared height in meters. Age was calculated for the time of surgery or visit date.
Genotyping
HuGE Navigator [16], a continuously updated database in human genome epidemiology, indexes all of the genetic association studies for a given disease or trait. There were 231 genes reported to be associated with OA as of June 2013. Due to limited budget, we chose to replicate all the SNPs that were reported in OA GWAS studies plus some SNPs from candidate studies.
Blood samples were obtained from all study participants and DNA was extracted by using the standard protocol. All the OA cases were genotyped by Sequenom iPLEX Gold method [17]. Briefly, 384-well plate chip on Sequenom using mass spectrometry was used for genotyping. Each multiplex PCR was done using 30ng of DNA (n=1: 1.25X PCR buffer Roche, 2mM MgCl2 Roche, 0.5M dNTPs, 0.11uM PCR primer pool oligos ordered from IDT, 0.15U/uL Roche FastStart) and the amplification was done following this cycling protocol: [95c 15min, 45x (95c 20sec, 58c 30sec, 72c 60sec), 72c 3min]. The SAP reaction was done to clean the PCR product and is followed by the extension reaction. After extension, the Salt Adduct Removal Step was run using 6mg of resin. The product was then spot on a Sequenom 384-well chip using a Nanodispenser, and loaded onto the Mass Spectrometer for reading.
All the controls were previously genotyped using Illumina HumanHap550-Duo BeadChip at Centrillion Biosciences at Palo Alto, California.
Cross-validation of genotyping quality was carried out on 31 controls that were genotyped by both Sequenom iPLEX Gold method and IlluminaHumanHap550-Duo BeadChip.
Statistical analysis
Distribution of age, gender, and BMI was examined and tested between OA cases and controls by either Chi-squared test or Student t-test wherever appropriate. Genotyping accuracy between two genotyping methods was evaluated by calculating genotype concordance rate in those subjects who were genotyped by both methods. Hardy-Weinberg Equilibrium (HWE) test was performed for each of the SNPs by exact Chi-squared test and removed in the subsequent analysis if p value <0.05. Chi-squared test was utilized to test the association between each of the SNPs and OA and logistic regression modeling was used to adjust for potential confounders including age, sex, and BMI. Significance level was set at alpha level of 0.0007 after correcting multiple testing with Bonferroni method. All analyses were done using STATA/SE 11.2 (Stata Corp, College Station, Texas, USA) except for HWE test which was performed by PLINK version 1.07 [18].
105 SNPs located in 71 genes were genotyped by Sequenom iPLEX Gold method for all cases and 52 healthy controls. 69 of these 105 SNPs were included in Illumina HumanHap550-Duo BeadChip and genotyped for all controls and therefore the subsequent analysis was focused on these 69 SNPs. 31 healthy controls were genotyped by both methods and were used to cross-validate the genotyping quality and accuracy between two genotyping methods. The genotype concordance rates for all 69 SNPs were 100%, indicating the comparability and accuracy of the two genotyping methods.Two SNPs were deviated from HWE and excluded from the subsequent analyses. Table 2 presents the results of the univariable and multivariable analysis of the association of each of the SNPs between OA and healthy controls. We found that minor alleles of rs2294995 located in COL9A3 and rs1049007 located in BMP2 were associated with one-third reduced risk for OA, but the significance did not survive the Bonferroni correction for multiple testing.
In the joint specific analyses, three SNPs were associated with knee OA [COL9A3 rs2294995 (OR:0.51, 95%CI: 0.32-0.8, P=0.004), HFE rs1799945 (OR:1.67, 95%CI: 1.06-2.6, P=0.025), and PACE4 rs900414 (OR:0.63, 95%CI: 0.41-0.97, P=0.036)], and five SNPs with hip OA [EDG2 rs10980705 (OR:0.4, 95%CI: 0.19-0.89, P=0.024), IL1RN rs315952 (OR:0.5, 95%CI: 0.26-0.93, P=0.03), BMP2 rs1049007 (OR:0.56, 95%CI: 0.33-0.95, P=0.03), IL1RN rs9005 (OR:1.69, 95%CI: 1.03-2.78, P=0.038), and COX2 rs5277 (OR:1.74, 95%CI: 1-3.01, P=0.048)], but all the significance did not survive the Bonferroni multiple testing correction.
Results
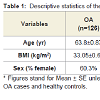
A total of 126 OA cases and 348 controls were included in this study. Among OA cases, 50 were males and 76 were females, 42 had total hip replacement and 84 had total knee replacement due to primary OA. Controls (146 males and 202 females) are healthy subjects who did not have evidence of joint OA. Characteristics of the study participants are presented in Table 1. OA cases were significantly older than healthy controls, and had a slightly higher BMI, which was not significant.105 SNPs located in 71 genes were genotyped by Sequenom iPLEX Gold method for all cases and 52 healthy controls. 69 of these 105 SNPs were included in Illumina HumanHap550-Duo BeadChip and genotyped for all controls and therefore the subsequent analysis was focused on these 69 SNPs. 31 healthy controls were genotyped by both methods and were used to cross-validate the genotyping quality and accuracy between two genotyping methods. The genotype concordance rates for all 69 SNPs were 100%, indicating the comparability and accuracy of the two genotyping methods.Two SNPs were deviated from HWE and excluded from the subsequent analyses. Table 2 presents the results of the univariable and multivariable analysis of the association of each of the SNPs between OA and healthy controls. We found that minor alleles of rs2294995 located in COL9A3 and rs1049007 located in BMP2 were associated with one-third reduced risk for OA, but the significance did not survive the Bonferroni correction for multiple testing.
In the joint specific analyses, three SNPs were associated with knee OA [COL9A3 rs2294995 (OR:0.51, 95%CI: 0.32-0.8, P=0.004), HFE rs1799945 (OR:1.67, 95%CI: 1.06-2.6, P=0.025), and PACE4 rs900414 (OR:0.63, 95%CI: 0.41-0.97, P=0.036)], and five SNPs with hip OA [EDG2 rs10980705 (OR:0.4, 95%CI: 0.19-0.89, P=0.024), IL1RN rs315952 (OR:0.5, 95%CI: 0.26-0.93, P=0.03), BMP2 rs1049007 (OR:0.56, 95%CI: 0.33-0.95, P=0.03), IL1RN rs9005 (OR:1.69, 95%CI: 1.03-2.78, P=0.038), and COX2 rs5277 (OR:1.74, 95%CI: 1-3.01, P=0.048)], but all the significance did not survive the Bonferroni multiple testing correction.
Discussion
The study is one of the few efforts on the replication of the previously identified genetic variants in OA. Advantages of the current study include using a genetically homogeneous population and advanced OA cases. The reduced genetic heterogeneity and using advanced OA cases are believed to increase study power in genetic association test.Although we were unable to replicate any of the SNPs that were previously reported to be associated with OA with a stringent significance level, two SNPs showed a potential to be associated with OA. One is rs2292995 located in COL9A3, which is a structure gene of articular cartilage. The gene has been associated with multiple epiphyseal dysplasia type 3 [19], and primary OA [20]. The second SNP is rs1049007 located in BMP2, which is involved in TGF-β signalling pathway that has been implicated in OA [21]. Although the associations for these SNPs did not reach the Bonferroni corrected significance level, it warrants further investigation in a large sample.
SNP rs143383 located in GDF5 gene is by far the most replicated SNP that is associated with OA. It was initially discovered in Asian population [22], followed by replication studies in European populations [23], and several meta-analyses confirmation [24,25]. The SNP was not included in the Illumina GWAS genotyping platform we used but we have a proxy SNP rs224329 which has r2=0.92 with rs143383. However, we could not detect any association between rs224329 and OA. Small sample size in the current study might be a possible explanation, but the results are the same as in the recent large GWAS on OA performed in the UK population [26], in which over 7410 OA cases and 11,009 unrelated population controls were included. Given the genetic similarity between Newfoundland and Labrador population and British population, the results may suggest the GDF5-OA association is population specific.
Data on replication of previously reported OA-associated genes are limited. GOAL study [27] utilized large number of symptomatic radiographic knee or hip OA and controls to replicate 68 variants in 12 genes including IL1A, IL1B, IL1RN, IL4R, IL6, COL2A1, ADAM12, ASPN, IGF1, TGFB1, ESR1 and VDR, but they did not replicate any of these associations. All the genetic variants they studied were included in our study. Similarly, these SNPs were not associated with OA in our sample; neither did in the large genome-wide meta-analysis [28].
Failure in the replication of previously reported genetic associations is common to the point that only less than 5% of the reported associations could be replicated in an independent study [29], which along with other factors has raised criticism on the usefulness of association studies [30]. The reasons for the failures are numerous: initially reported associations might be false positives, or replications might be false negative findings resulting from a biased sampling, hidden or uncorrected population stratification, small sample sizes. The association might only hold true for the population in which the association was reported due to the differences and specificity of LD patterns across different populations. The complexity of multifactorial traits is another issue: the effect of multiple genetic variants and heterogeneity means that the presence of all of the risk alleles together may not be required for the disease to develop; therefore, distribution of risk alleles in two groups of cases from different or even same population might be different from each other. All these factors are aggravated in the study of OA due to the age dependency and the lack of a unique method for the ascertainment of the study subjects used by different investigators.
The above-mentioned reasons could partially explain why we were not able to replicate the majority of the SNPs in our study. In addition, our study has been limited by a number of factors: sample size is relatively small. Given our sample size and assuming minor allele frequency of 35% in controls, we have 80% power to only detect an OR of 1.8 or above at an alpha level of 0.05. The minimum detectable OR would be 2.5 if we define the significance level at 0.0007 after taking into account of multiple testing. However, the study power is maximized by the optimal case-control ratio and using extreme severe OA cases. We recently found that NL population has slightly subtle population stratification (not published data), which might lead to false negatives.
In conclusion, we were unable to replicate the previously reported OA-associated genes, but two SNPs showed suggestive associations with plausible biological mechanism. Further studies are needed to confirm the results.
Acknowledgements
We thank all the study participants who made the study possible. The staff from Quebec Genome and Innovation Centre and Centrillion Biosciences, CA, were acknowledged for their help in genotyping.References
- Kean WF, Kean R, Buchanan WW (2004) Osteoarthritis: symptoms, signs and source of pain. Inflammopharmacology 12: 3-31.
- Guccione AA, Felson DT, Anderson JJ, Anthony JM, Zhang Y, et al. (1994) The effects of specific medical conditions on the functional limitations of elders in the Framingham Study. Am J Public Health 84: 351-358.
- Arden N, Nevitt MC (2006) Osteoarthritis: Epidemiology. Best Pract Res Clin Rheumatol 20: 3-25.
- Maetzel A, Li LC, Pencharz J, Tomlinson G, Bombardier C (2004) The economic burden associated with osteoarthritis, rheumatoid arthritis, and hypertension: A comparative study. Ann Rheum Dis 63: 395-401.
- (2007) National and State Medical Expenditures and Lost Earnings Attributable to Arthritis and Other Rheumatic Conditions---United States, 2003. MMWR 56: 4-7.
- Felson DT (2004) An update on the pathogenesis and epidemiology of osteoarthritis. Radiol Clin North Am 42: 1-9.
- Hunter DJ, March L, Sambrook PN (2002) Knee osteoarthritis: the influence of environmental factors. Clin Exp Rheumatol 20: 93-100.
- MacGregor AJ, Antoniades L, Matson M, Andrew T, Spector TD (2000) The genetic contribution to radiographic hip osteoarthritis in women: results of a classic twin study. Arthritis Rheum 43: 2410-2416.
- Spector TD, Cicuttini F, Baker J, Loughlin J, Hart D (1996) Genetic influences on osteoarthritis in women: a twin study. BMJ 312: 940-944.
- Hirsch R, Lethbridge-Cejku M, Hanson R, Scott WW Jr, Reichle R, et al. (1998) Familial aggregation of osteoarthritis: data from the Baltimore Longitudinal Study on Aging. Arthritis Rheum 4: 1227-1232.
- Riyazi N, Meulenbelt I, Kroon HM, Ronday KH, Hellio le Graverand MP, et al. (2005) Evidence for familial aggregation of hand, hip, and spine but not knee osteoarthritis in siblings with multiple joint involvement: the GARP study. Ann Rheum Dis 64: 438-443.
- Lafeber FP, van Spil WE (2013) Osteoarthritis year 2013 in review: biomarkers; reflecting before moving forward, one step at a time, Osteoarthritis Cartilage 21: 1452-1464.
- Heutink P, Oostra BA (2002) Gene finding in genetically isolated populations. Hum Mol Genet 11: 2507-2015.
- Rahman P, Jones A, Curtis J, Bartlett S, Peddle L, et al. (2003) The Newfoundland population: a unique resource for genetic investigation of complex diseases. Hum Mol Genet 13: 1287.
- Kristiansson K, Naukkarinen J, Peltonen L (2008) Isolated populations and complex disease gene identification. Genome Biol 9: 109.
- Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ (2008) A Navigator for Human Genome Epidemiology. Nat Genet 40: 124-125.
- Gabriel S, Ziaugra L, Tabbaa D (2009) SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet.
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 8: 559-575.
- Jackson GC, Marcus-Soekarman D, Stolte-Dijkstra I, Verrips A, Taylor JA, et al. (2010) Type IX collagen gene mutations can result in multiple epiphyseal dysplasia that is associated with osteochondritis dissecans and a mild myopathy. Am J Med Genet A. 152A: 863-869.
- Ikeda T, Mabuchi A, Fukuda A, Kawakami A, Ryo Y, et al. (2002) Association analysis of single nucleotide polymorphisms in cartilage-specific collagen genes with knee and hip osteoarthritis in the Japanese population. J Bone Miner Res 17: 1290-1296.
- Finnson KW, Chi Y, Bou-Gharios G, Leask A, Philip A (2012) TGF-b signaling in cartilage homeostasis and osteoarthritis. Front Biosci (Schol Ed) 4: 251-268.
- Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, et al. (2007) A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet 39: 529-533.
- Southam L, Rodriguez-Lopez J, Wilkins JM, Pombo-Suarez M, Snelling S, et al. (2007) An SNP in the 5′-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum Mol Genet 16: 2226-2232.
- Liu J, Cai W, Zhang H, He C, Deng L (2013) Rs143383 in the growth differentiation factor 5 (GDF5) gene significantly associated with osteoarthritis (OA)-a comprehensive meta-analysis. Int J Med Sci 10: 312-319.
- Hao SW, Jin QH (2013) Association between the +104T/C polymorphism in the 5'UTR of GDF5 and susceptibility to knee osteoarthritis: a meta-analysis. Mol Med Rep 7: 485-488.
- Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, Day-Williams AG, et al. (2012) Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 380: 815-823.
- Limer KL, Tosh K, Bujac SR, McConnell R, Doherty S, et al. (2009) Attempt to replicate published genetic associations in a large, well-defined osteoarthritis case-control population (the GOAL study). Osteoarthritis Cartilage 17: 782-789.
- Evangelou E1, Kerkhof HJ, Styrkarsdottir U, Ntzani EE, Bos SD, et al. (2013) A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann Rheum Dis.
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K (2002) A comprehensive review of genetic association studies. Genet Med 4: 45-61.
- Couzin-Frankel J (2010) Major heart disease genes prove elusive. Science 328: 1220-1221.