Journal of Forensic Investigation
DNA Microarray Analysis of Hypothermic Murine Myocardium to Study Pathophysiology and Identify Forensic Biomarkers
Masataka Takamiya1*, Kiyoshi Saigusa2 and Koji Dewa1
- 1Department of Forensic Medicine, Iwate Medical University, Iwate, Japan
- 2Department of Biology, Iwate Medical University, Iwate, Japan
*Address for Correspondence: Masataka Takamiya MD, PhD, Department of Forensic Medicine, Iwate Medical University, 2-1-1 Nishitokuta, Yahaba, Iwate 028-3694, Japan, Tel: +81-19-698-1820; Fax: +81-19-908-8005; E-mail: mtakamiy@iwate-med.ac.jp
Citation: Takamiya M, Saigusa K, Dewa K. DNA Microarray Analysis of Hypothermic Murine Myocardium to Study Pathophysiology and Identify Forensic Biomarkers. J Forensic Investigation. 2016; 4(1): 11.
Copyright © 2016 Takamiya M, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Forensic Investigation | ISSN: 2330-0396 | Volume: 4, Issue: 1
Submission: 20 January, 2016 | Accepted: 16 March, 2016 | Published: 21 March, 2016
Abstract
We used DNA microarray technology to analyze the myocardial transcriptome of mice killed via experimentally induced hypothermia. This analysis identified significant differential regulation of 3438 genes; specifically, 1704 genes were upregulated, and 1734 were downregulated in response to hypothermia. The gene encoding granzyme A was the most upregulated gene, and that encoding solute carrier family 41 member 3 was the most downregulated. Gene-set analysis identified significant hypothermia-induced variation in 79 pathways, and we suggest that pathways related to granzyme A and cell death may be involved in cardiac pathogenesis of hypothermia. Gene-function-category analysis demonstrated the most highly represented categories among the upregulated and downregulated genes were cellular process (biological process), binding (molecular function), and cell and cell part (cellular component). The presented findings clearly demonstrated that acute myocardial responses to hypothermia did occur; they also indicated several cardiac-related candidate genes as forensic biomarkers of hypothermia. Hypothermia-induced myocardial cell death would be an irreversible change that, we believe, may explain the circulatory failure, resistance to treatment, and high mortality associated with hypothermia. Furthermore, the present microarray data may facilitate development of immunohistochemical analysis and protocols to be used for human forensics and may be beneficial in clinical research on hypothermia.
Keywords
Heart; Myocardium; Hypothermia; Transcriptome; DNA microarray; Quantitative PCR
Introduction
Hypothermia is classically defined as a core body temperature of less than 35 °C. Hypothermia develops when adaptive thermoregulatory mechanisms are overwhelmed and hypothermia is a common danger even indoors and in temperate climates [1,2]. Macroscopically, hypothermia may cause the following: frostbite; bright-pink lividity [3]; hemorrhages in muscles [4]; cerebral edema [3]; venous thrombosis [2]; pulmonary edema [2,3, 5]; bronchopneumonia [6]; hemorrhages in the stomach, ileum, and colon [3,5,6]; and diuresis [5]. Additionally, histological examinations reveal fatty changes in the liver and kidneys [7-9], vacuolization of liver cells [3] and pancreatic adenoid cells [10], renal tubular necrosis [5,6], heat shock protein 70 accumulation in renal tubular epithelium and glomerular podocytes [11], and hemorrhagic pancreatitis [3,5,6,8]. Forensic differential diagnosis of hypothermia is often difficult because many of these autopsy findings are not specific to hypothermia. Therefore, diagnosis of hypothermia must be based partly on exclusion criteria and historical information. However, molecular biological methods may provide more definitive criteria for forensic diagnosis of hypothermia [12].
Hypothermia carries a high mortality rate due to circulatory failure [13]. Cardiac output and sinus rate decrease and arrhythmias (from atrial fibrillation to ventricular fibrillation) occur during hypothermia [14,15]. Therefore, it is important to document the cardiovascular state during hypothermia; consequently, we considered it worthwhile to assess hypothermia-induced changes to the myocardial transcriptome. In this study, the myocardial transcriptome in hypothermic mice was analyzed to examine the cardiac pathophysiology of mammalian hypothermia and to identify candidates for forensic biomarkers of human hypothermia. This study was designed both for the study of cardiac pathophysiology and to improve forensic practices; moreover, the findings should be beneficial for clinical research on hypothermia.
Materials and Methods
Tissue samples
A water-bath method described previously was adapted to induce hypothermia in mice [16]. Male ddY mice 7 weeks of age and weighing 36.3 ± 6.8 g were housed under controlled lighting (lights on at 7:00 am and off at 7:00 pm) and given free access to food and water. This was a preliminary study, and only male mice were used; therefore, potential differences due to sex should be addressed in future studies. Each mouse was anesthetized by sevoflurane inhalation and then confined in a metallic restraint cage that was kept in a water bath set at 10 °C such that each mouse was immersed up to the neck in the cold water. Each animal died from continuous exposure to cold water for 42.8 ± 12.6 minutes. Immediately after death, a 2-mm thick specimen from the anterior wall of the left ventricle was resected 3 mm from the heart apex for each animal. In all, 28 mice were subjected to hypothermia-induced death; four were used for DNA microarray analyses, four for selection of genes to be used as the internal standard, 10 for quantitative PCR analyses, and 10 for immunohistochemical analyses. Control mice (n = 28) were sacrificed by inhalation of CO2, and the same part of each control left ventricle was examined (n = 4 for DNA microarray, n = 4 for internal standard gene selection, n = 10 for quantitative PCR, n = 10 for immunohistochemistry). Because this was a forensic pathologic study, samples from hypothermic and separately control mice were collected from cadavers. In other words, death was the most important commonality between the hypothermic and control mice. Furthermore, the interval between initial CO2 exposure and death was extremely short in the control group, and this short time period may have precluded substantial changes in gene expression; therefore, CO2 exposure was an appropriate negativecontrol treatment for our purposes. For DNA microarray analyses, internal standard gene selection, and quantitative PCR analyses, each isolated tissue specimen was immediately soaked in 1.5 ml RNAlater solution (Applied Biosystems, Carlsbad, CA), and stored at -80 °C for 2 weeks. The research described in this report was conducted in accordance with the guidelines for animal experimentation from Iwate Medical University.
DNA microarray methods
RNeasy Fibrous Tissue Mini Kits (Qiagen, Valencia, CA) were used to extract total RNA samples from tissue specimens. All microarray procedures described here were performed at Tohoku Chemical Research Institute of Bio-system Informatics (Morioka, Japan). Electrophoresis through a 1% agarose gel, an absorptiometer, and a 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA) were used to confirm the quality of the RNA samples; quality control criteria were as follows: OD260/280 > 1.5, OD260/230 > 1.0, no degradation in electrophoresis, 28S/18S ribosomal RNA bands > 1.8, RNA Integrity Number > 7. The Mouse GE 4x44K v2 Microarray Kit (4 arrays, Agilent Technologies, Santa Clara, CA) was used according to the manufacturer’s instructions to determine gene expression profiles. The analysis was performed with the two-color method; control samples were labeled with Cy3, and hypothermic samples were labeled with Cy5. Scanning of microarray slides was performed with Agilent Technologies Scanner G2505C and Agilent Feature Extraction 10.7.3.1 (Agilent Technologies, Santa Clara, CA). For further analysis, GeneSpring (Agilent Technologies, Santa Clara, CA) was also used. In addition, annotations of genes were based on the Gene and GenBank of National Center for Biotechnology Information (NCBI, Bethesda, MD). Thus, those probes that are not cataloged in Gene or GenBank were not annotated in the present study. This experiment was verified according to the minimum information about microarray experiment (MIAME) guidelines (Supplement 1) [17].
Quality control: Quality control for each feature (spot) was performed using the settings recommended by Agilent Technologies. Background signal was subtracted, and signal intensity of each feature was globally normalized via locally weighted scatter-plot smoothing (LOWESS). The following flag parameters were used: “Feature is saturated”; “Feature is not uniform”; “Feature is not positive and significant”; “Feature is not above background”; “Feature is a population outlier”. For each of these parameters, one of thefollowing terms was applied to each feature:
Detected: The data are reliable.
Not detected: The quality of data are undetermined.
Compromised: The data are unreliable.
Each gene was represented by four spots, and one of the following terms was applied to each spot:
Detected: All the parameters are “Detected”.
Not detected: The parameters are combinations of “Detected” and “Not detected”.
Compromised: One of the parameters is “Compromised”.
For subsequent analyses, we used only genes for which all four spots were categorized as “Detected”.
Selection of significantly regulated genes: The one-sample Student’s t-test was performed to identify genes that exhibited significant differential expression in response to hypothermia. P values of 0.05 or lower were considered statistically significant. After the correction for multiple tests (Benjamini-Hochberg method), only five genes showed significant differences between the experimental and control samples. This phenomenon was due to the dispersion of gene expression data among four arrays. Therefore, these statistical methods were not used.
Gene-set analysis:
KEGG analysis: We used the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database and previously published methods to perform gene-set analysis [18]. The mean of fold-changes among all quality-controlled genes were compared to the fold-change of each gene set. Z scores of fold-changes were calculated, and datasets with normal distributions were subject to statistical analysis. P values of 0.05 or lower were considered statistically significant.
Analyses with publically available pathway databases: Biocarta pathway (San Diego, CA) and GeneAssist (Applied Biosystems, Waltham, MA) are publically available pathway databases. In addition to KEGG, supplementary analyses of gene sets were performed with each of these databases. For pathway selection, three upregulated genes (granzyme A: Gzma; cytochrome P450, family 26, subfamily b, polypeptide 1: Cyp26b1; and activating transcription factor 3: Atf3) and three downregulated genes (solute carrier family 41, member 3: Slc41a3; solute carrier family 46, member 2: Slc46a2; and nuclear receptor subfamily 1, group D, member 1: Nr1d1) were used.
Analysis with Biocarta pathway: Biocarta pathway (San Diego, CA) is an open-source database; the information is displayed in a graphical format, and known genomic and proteomic relationships are mapped. In connection with the six selected genes, a granzyme A-mediated apoptosis pathway was retrieved for Gzma. Using previously published methods, a gene-set analysis was performed with this pathway [18].
Analysis with GeneAssist: More than 350 different pathways are currently available in GeneAssist (Applied Biosystems, Waltham, MA). Illustrations show molecular interactions and cellular compartments. In connection with the six selected genes, the following five pathways were found in this database: the granzyme, granzyme A, IL 9 pathways (for Gzma), and the MAPK signaling and TNF signaling pathways (for Atf3). Using previously published methods, gene-set analyses were performed with these pathways [18].
Gene functional-category analysis: Gene functional-category analyses were performed. The number of genes corresponding to each Gene Ontology term among all genes was compared to the number among differentially regulated genes using Fisher’s Exact Test. P values of 0.05 or lower was considered statistically significant.
Quantitative PCR
To validate selected aspects of microarray results, three upregulated genes (Gzma, Cyp26b1, Atf3; Table 1) and three downregulated genes (Slc41a3, Slc46a2, Nr1d1; Table 2) were selected for measurement by quantitative PCR.
RNA extraction and reverse transcription: Total RNA was extracted from tissues using an RNeasy Fibrous Tissue Mini Kit (Qiagen, Valencia, CA). An absorptiometer and electrophoresis through a 1% agarose gel were used to confirm the quality of the RNA samples; the quality control criteria were as follows: OD260/280 > 1.5, OD260/230 > 1.0, no degradation in electrophoresis with 28S and 18S ribosomal RNA bands. The RNA was treated with TURBO DNase (Applied Biosystems, Waltham, MA). A High-Capacity cDNA Reverse Transcription kit with RNase inhibitor (Applied Biosystems, Waltham, MA) was then used according to the manufacturer’s instructions to synthesize cDNA.
Real-time quantitative PCR: TaqMan Gene Expression Assays (Applied Biosystems, Waltham, MA) are the most reliable sets of predesigned quantitative real-time PCR assays. TaqMan Gene Expression Assays (Applied Biosystems, Waltham, MA) have been designed using the validated bioinformatics pipeline and run with the same PCR protocol; this protocol eliminates the need for time-consuming primer design or PCR optimization. TaqMan Gene Expression Assays (Applied Biosystems, Waltham, MA) are used as the gold standard technology in mRNA quantification. In the present study, TaqMan Gene Expression Assays (Applied Biosystems, Waltham, MA) were used with the following primers and probes:
Gzma: Mm01304452_m1, NM_010370.2, 59bp
Cyp26b1: Mm00558507_m1, NM_001177713.1, NM_175475.3, 75bp
Atf3: Mm00476032_m1, NM_007498.3, 61bp
Slc41a3: Mm01182529_m1, NM_001037493.2, NM_027868.2, 91bp
Slc46a2: Mm00498614_m1, NM_021053.4, 64bp
Nr1d1: Mm00520708_m1, NM_145434.4, 62bp
A value that represents the stability of individual internal standard genes was used to select the internal-standard gene; values for β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and 18S ribosomal RNA were 1.68, 1.52 and 1.96 respectively [19]. Therefore, GAPDH was used as the internal-standard gene. GAPDH primers and probes are supplied with TaqMan Gene Expression Assays (Mm99999915_g1, NM_001289726.1, NM_008084.3, 107bp, Applied Biosystems, Waltham, MA). Real-time quantitative PCR was performed using a PRISM 7500 sequence detector (Applied Biosystems, Waltham, MA). Individual 50-μl reaction mixtures containing TaqMan Gene Expression Master Mix (Applied Biosystems, Waltham, MA) and the thermal cycler conditions recommended in the manufacturer’s instructions were used. All samples were analyzed in triplicate. No amplification was evident in any no-template control. The relative expression of each target gene was calculated via the ΔΔ Ct method as described in the manufacturer’s instructions (Applied Biosystems, Waltham, MA). Differences in gene expression between control and hypothermic myocardium were assessed via the Student’s t test; P values of 0.05 or lower were considered statistically significant. In addition, this experiment was checked according to the quantitative real-time PCR experiment (MIQE) guidelines (Supplement 2) [20].
Immunohistochemistry
Specimens were fixed in 4% buffered formalin, embedded in paraffin, and sectioned at a thickness of 2.5 μm. Each section was stained with hematoxylin and eosin (H&E). In order to analyze histological dynamics, immunohistochemistry was performed. No commercially suitable antibodies were found for two upregulated genes (Cyp26b1, Atf3) or three downregulated genes (Slc41a3, Slc46a2, Nr1d1). Therefore, expression of granzyme A was examined using rabbit anti-granzyme A (Cloud-Clone, Houston, TX) as the primary antibodies. Antigen activation was performed with a microwave oven and antigen activating solution (Nichirei, Tokyo, Japan). Sections were incubated with primary antibodies diluted 1:25 for 12 h in a humid chamber at 4 °C. Thereafter, sections were incubated with peroxidase-conjugated secondary antibody according to the manufacturer’s instructions (Histofine MAX-PO (R), Nichirei, Tokyo, Japan). The chromogen used to visualize antibody signal was 3, 3-diamino-benzidine (Nichirei, Tokyo, Japan), and the specimens were counterstained with hematoxylin. Quantitative analysis was difficult in cardiac tissue; therefore, expression was assessed qualitatively via light microscope.
Results
DNA microarrays
Quality control: With the quality control measures, data from 17870 of the 39485 microarray genes could be used for further analysis.
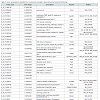
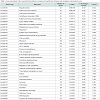
Identification of significantly regulated genes: We identified 3438 genes that exhibited hypothermia-induced differential expression in mouse myocardium; 1704 were upregulated, and 1734 were downregulated. Of the upregulated genes, 120 genes increased by > 1.5-fold and ≤ 2-fold, five by > 2-fold and ≤ 3-fold, and three by > 3-fold. Among the genes confirmed as upregulated, granzyme A was the most upregulated gene with a 3.26-fold increase. For the downregulated genes, expression in hypothermic myocardium was lower than in control myocardium by < 0.6-fold and ≥ 0.5-fold for 22 genes, by < 0.5-fold and ≥ 0.4-fold for seven genes, and by < 0.4- fold for one gene. Solute carrier family 41 member 3 was the most downregulated gene with a 0.463-fold reduction. Upregulated genes and downregulated genes were arranged in order (descending or ascending, respectively) with respect to fold-change; Table 1 lists the 50 upregulated genes with a change greater than 1.6-fold, and Table 2 lists the 30 downregulated genes with a change of 0.6-fold or lower.
Gene-set analysis:
KEGG analysis: Significant variations were found in 79 pathways, and the gene sets that were upregulated or downregulated are summarized in Tables 3 and 4 respectively. A total of 57 pathways were significantly upregulated, and 22 pathways were downregulated. Upregulation of the viral myocarditis pathway was evident. But, no pathway related to granzyme A and/or activating transcription factor 3 was identified as significantly altered in hypothermic myocardium.
Publically available database analysis:
Analysis with Biocarta pathway: In connection with the six selected genes (Gzma, Cyp26b1, Atf3, Slc41a3, Slc46a2 and Nr1d1), a granzyme A-mediated apoptosis pathway was retrieved for Gzma. This pathway was significantly upregulated (Figure 1 and Table 5).
Analysis with GeneAssist: In connection with the six selected genes, the following five pathways were found in this database: the granzyme, granzyme A, and IL 9 pathways for Gzma and the MAPK signaling and TNF signaling pathways for Atf3. Among these five pathways, the granzyme pathway, granzyme A pathway (for Gzma), and MAPK signaling (for Atf3) were significantly upregulated (Table 5).
Analysis of gene function category: To investigate the biological functions involving the differentially regulated genes, we performed Gene Ontology category analysis. The 10 categories most commonly associated with these differentially regulated (upregulated or downregulated) genes included biological process, molecular function, and cellular component. The most commonly represented categories among upregulated and downregulated genes were cellular process, binding, cell, and cell part (Supplements 3 and 4).
Validation of gene expression results by quantitative PCR
We used quantitative PCR to validate microarray findings for three upregulated and three downregulated genes (Figure 2). Quantitative PCR findings were consistent with DNA microarray findings except that fold differences detected by the two methods were not identical for each gene. Among the downregulated genes, Slc41a3 was decreased to a greater extent than Slc46a2 or Nr1d1 was.
Immunohistochemical analyses
Myocardium remained stable based on H&E-stained specimens. No infiltration of inflammatory cells was seen in hypothermic specimens. Based on the immunohistochemical analyses, granzyme A expression was not evident in any control myocardium specimen; in contrast, slight granzyme A expression was evident in cytoplasm of hypothermic cardiac myocytes. There was no granzyme A expression in endocardial, epicardial, or endothelial cells (Figure 3).
Discussion
In the present study, DNA microarray analyses were performed with control and hypothermic ventricular myocardium. A total of 3438 genes were found to be differentially expressed in hypothermic tissue; specifically, 1704 were upregulated, and 1734 were downregulated. Cardiac candidates for forensic biomarkers of hypothermia were Gzma, Cyp26b1, and Atf3 among the upregulated genes (Table 1) and Slc41a3, Slc46a2, and Nr1d1 among the downregulated genes (Table 2). Needless to say, the possibility exists that other currently unidentified factors that are differentially expressed may be useful for diagnosing hypothermia. To our knowledge, no previous studies of hypothermia have identified a connection between hypothermia and Gzma, Cyp26b1, Atf3, Slc41a3, Slc46a2 or Nr1d1 expression. The Gene Ontology analysis indicated that cellular process, binding, cell, and cell part were significantly highly represented categories. The gene-set analysis revealed that cell death pathways related to granzyme A were upregulated and that these pathways may be involved in the cardiac pathogenesis of hypothermia.
Multiple types of cell death have been described; these include apoptosis, autophagy, necrosis, senescence, and mitotic catastrophe [21]. Some genes identified here as differentially regulated in response to hypothermia are modulators of one or more cell-death pathways; for example, granzyme A was upregulated in this study, and this protein leads to cell death by mediating cleavage of DNA molecules in the nucleus and of molecules in the nuclear envelope or mitochondrial membranes [21,22]. In addition, granzyme A plays an important role in inflammation [21]. Extracellular matrix components such as vitronectin, fibronectin, fibrinogen, laminin, and proteoglycans can be cleaved by granzyme A and this degradation may facilitate movements of leukocytes [21,23]. Granzyme A can also induce secretion of cytokines [24-26]. Activating transcription factor 3 was also upregulated in the hypothermic myocardium, and it is known to affect genes involved in apoptosis [27].
Decreased cardiac output and fatal arrhythmias are observed during hypothermia [14,15]. Histological analyses reveal that hypothermia causes degenerative foci in myocardium [28]; these foci consist of myofibrosis, calcifications, hyperchromasia, contraction bands, waving, discoloration and fragmentation of the fibers, edema, hemorrhages [3,29], coagulation necrosis, extravasation of red cells [3], inflammatory reactions (including cellular infiltration), granulation, and scarring [28,30-31]. Fatty changes in myocardium are also observed [32]. These histological lesions could act as foci of ventricular fibrillation [31]. Putative causative mechanisms include myocardial ischemia [33] mediated by platelet thrombi [31], increased blood viscosity [29,34-36], insufficient coronary perfusion, and catecholamine mobilizations [29]. Interestingly, our current findings suggested a previously unidentified putative mechanism for these lesions; specifically, hypothermia may induce myocardial cell death via granzyme A and activating transcription factor 3, and this cell death may cause some of the histological lesions and clinical symptoms characteristic of hypothermic myocardium. Some clinical studies show that hypothermia causes high mortality [13]. Myocardial cell deaths induced by hypothermia would be irreversible changes, and we postulate that this cell death may be related to the circulatory failure, resistance to treatment, and high mortality associated with hypothermia.
A compact arrangement of myocardial fibers (called interstitial narrowness) between myocardial fibers has recently been identified as a histological finding indicative of hypothermia [37]. Some pathologists have proposed that this finding is caused by dehydration during hypothermia [38]. The present transcriptome analysis did not provide substantial insight into this mechanism and related genetic pathways responsible for this phenomenon. One might say that interstitial narrowness is a passive myocardial change caused by dehydration.
The present research demonstrated changes in gene expression in hypothermic murine myocardium and identified potential cardiac candidates for forensic biomarkers of hypothermia. The present findings suggested that myocardial cell death was induced in hypothermia and that this cell death could cause cardiac dysfunctions and high mortality during hypothermia. These data may help elucidate the pathophysiology of hypothermia. But mechanisms of hypothermia may be multifactorial and manifold. In addition, reports of forensic hypothermia investigations that use molecular biological techniques have only recently begun to appear, and traditional characteristics of hypothermia remain important for forensic diagnosis [39]. Large-scale gene analyses, including DNA microarray analyses, are promising methods for detecting forensic pathological biomarkers. However, it should be noted that applications of these gene expression data would be limited in routine forensic practices. For example, the postmortem interval could negatively affect gene expression analysis. The samples in these animal experiments were collected under ideal circumstances. After considering these transcriptomic data along with postmortem changes, expression of biomarker candidates in forensic human samples should be assessed. Because RNA decomposes in the postmortem interval, utilization of protein markers should be considered as alternative to RNA biomarkers. Therefore, analysis of the proteins encoded by candidate biomarkers identified via this DNA microarray analysis is a next step in the process of identifying clinically useful biomarkers. The present histological study also demonstrated slight expression of granzyme A in cardiac myocytes. Importantly, results of the present murine DNA microarray study may provide data applicable to the development of future immunohistochemical protocols for analysis of human forensic samples. In addition, we believe that these data are informative, not only for future forensic pathological studies, but also potentially for clinical research into hypothermia.
Conclusion
We compared the hypothermic murine myocardial transcriptome to that of a room-temperature control using DNA microarray technology; we thereby identified 3438 genes that were significantly differentially expressed in response to hypothermia. Of these genes, 1704 were upregulated and 1734 were downregulated. The gene encoding granzyme A was the most upregulated gene, and that encoding solute carrier family 41 member 3 was the most downregulated. In our gene-set analysis, significant variations were found in 79 pathways, and we suggest that pathways related to granzyme A and cell death may be involved in hypothermia-induced cardiac pathogenesis. The present study documented acute myocardial responses during hypothermia and revealed cardiac candidates that may be useful as forensic biomarkers of hypothermia. Myocardial cell death induced by granzyme A may represent an irreversible change that is related to the circulatory failure, resistance to treatment, and high mortality associated with hypothermia. Furthermore, the present microarray data may facilitate development of protocols for immunohistochemical analysis of specimens from human cases and be beneficial to clinical research on hypothermia.
Acknowledgements
This study was supported by JSPS KAKENHI grant number 26460884.
References
- Ulrich AS, Rathlev NK (2004) Hypothermia and localized cold injuries. Emerg Med Clin North Am 22: 281-298.
- Saukko P, Knight B (2004) Neglect, starvation and hypothermia: Injury caused by cold: hypothermia. In: Saukko P, Knight B, (Eds) Knight’s Forensic Pathology (3rd Edn), Edward Arnold (Publishers) Ltd, CRC Press, London, pp. 414-420.
- Hirvonen J (1976) Necropsy findings in fatal hypothermia cases. Forensic Sci 8: 155-164.
- Aghayev E, Thali MJ, Jackowski C, Sonnenschein M, Dirnhofer R, et al. (2008) MRI detects hemorrhages in the muscles of the back in hypothermia. Forensic Sci Int 176: 183-186.
- Paton BC (1983) Accidental hypothermia. Pharmacol Ther 22: 331-377.
- DiMaio VJ, DiMaio D (2001) Hyperthermia and Hypothermia: The effects of heat and cold. In: DiMaio VJ, DiMaio D, (Eds) Forensic pathology (2nd Edn), Boca Raton, CRC press, pp. 419-434.
- Fisher ER, Fedor EJ, Fisher B (1957) Pathologic and histochemical observations in experimental hypothermia. AMA Arch Surg 75: 817-827.
- Mant AK (1969) Autopsy diagnosis of accidental hypothermia. J Forensic Med 16: 126-129.
- Preuss J, Dettmeyer R, Lignitz E, Madea B (2004) Fatty degeneration in renal tubule epithelium in accidental hypothermia victims. Forensic Sci Int 141: 131-135.
- Preuss J, Lignitz E, Dettmeyer R, Madea B (2007) Pancreatic changes in cases of death due to hypothermia. Forensic Sci Int 166: 194-198.
- Preuss J, Dettmeyer R, Poster S, Lignitz E, Madea B (2008) The expression of heat shock protein 70 in kidneys in cases of death due to hypothermia. Forensic Sci Int 176: 248-252.
- Hirvonen J, Huttunen P (1982) Increased urinary concentration of catecholamines in hypothermia deaths. J Forensic Sci 27: 264-271.
- Vassal T, Benoit-Gonin B, Carrat F, Guidet B, Maury E, et al. (2001) Severe accidental hypothermia treated in an ICU: prognosis and outcome. Chest 120: 1998-2003.
- Lauri T, Leskinen M, Timisjarvi J, Hirvonen L (1991) Cardiac function in hypothermia. Arctic Med Res 50 Suppl 6: 63-66.
- Maaravi Y, Weiss AT (1990) The effect of prolonged hypothermia on cardiac function in a young patient with accidental hypothermia. Chest 98: 1019-1020.
- Okuda C, Saito A, Miyazaki M, Kuriyama K (1986) Alteration of the turnover of dopamine and 5-hydroxytryptamine in rat brain associated with hypothermia. Pharmacol Biochem Behav 24: 79-83.
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, et al. (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29: 365-371.
- Kim SY, Volsky DJ (2005) PAGE: Parametric analysis of gene set enrichment. BMC Bioinformatics 6: 144.
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034.
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611-622.
- Zhou F (2010) Expression of multiple granzymes by cytotoxic T lymphocyte implies that they activate diverse apoptotic pathways in target cells. Int Rev Immunol 29: 38-55.
- Buzza MS, Bird PI (2006) Extracellular granzymes: current perspectives. Biol Chem 387: 827-837.
- Krishan K, Kanchan T, DiMaggio JA (2015) Emergence of forensic podiatry--a novel sub-discipline of forensic sciences. Forensic Sci Int 255: 16-27.
- Metkar SS, Menaa C, Pardo J, Wang B, Wallich R, et al. (2008) Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity 29: 720-733.
- Sower LE, Klimpel GR, Hanna W, Froelich CJ (1996) Extracellular activities of human granzymes. I. Granzyme A induces IL6 and IL8 production in fibroblast and epithelial cell lines. Cell Immunol 171: 159-163.
- Yoshikawa Y, Hirayasu H, Tsuzuki S, Fushiki T (2008) Granzyme A causes detachment of alveolar epithelial A549 cells accompanied by promotion of interleukin-8 release. Biosci Biotechnol Biochem 72: 2481-2484.
- Turchi L, Aberdam E, Mazure N, Pouyssegur J, Deckert M, et al. (2008) Hif-2alpha mediates UV-induced apoptosis through a novel ATF3-dependent death pathway. Cell Death Differ 15: 1472-1480.
- Hirvonen J, Huttunen P, Hiltunen K (1988) Creatine phosphokinase in serum and cerebrospinal fluid, and microscopic findings in brain and heart in hypothermic rabbits. Forensic Sci Int 39: 271-278.
- Hirvonen J, Penttinen J, Huttunen P, Saukko P (1980) Changes in the myocardium and skeletal muscle in guinea pigs in cold exposure with and without ethanol. Z Rechtsmed 84: 195-207.
- Altland PD, Highman B, Sellner RG (1974) Serum enzyme and tissue changes in shaven rabbits exposed to cold. Cryobiology 11: 296-304.
- Sarajas HS (1964) Myocardial damage induced by immersion hypothermia. Am J Cardiol 13: 355-366.
- Preuss J, Dettmeyer R, Lignitz E, Madea B (2006) Fatty degeneration of myocardial cells as a sign of death due to hypothermia versus degenerative deposition of lipofuscin. Forensic Sci Int 159: 1-5.
- Lynch HF, Adolph EF (1957) Blood flow in small blood vessels during deep hypothermia. J Appl Physiol 11: 192-196.
- Kanter GS (1968) Hypothermic hemoconcentration. Am J Physiol 214: 856-859.
- Keen G, Gerbode F (1963) Observations on the microcirculation during profound hypothermia. J Thorac Cardiovasc Surg 45: 252-260.
- Taylor MJ, Bailes JE, Elrifai AM, Shih SR, Teeple E, et al. (1995) A new solution for life without blood. Asanguineous low-flow perfusion of a whole-body perfusate during 3 hours of cardiac arrest and profound hypothermia. Circulation 91: 431-444.
- Funayama M, Morita M, Shimizu K, Shiono H, Hiraiwa K, et al. (1997) Compact arrangement of myocardial fibers in cases of fatal hypothermia. Proceeding of 14th meeting of International Association of Forensic Sciences. Curr Top Forensic Sci 3: 381-383.
- Jolly BT, Ghezzi KT (1992) Accidental hypothermia. Emerg Med Clin North Am 10: 311-327.
- Turk EE (2010) Hypothermia. Forensic Sci Med Pathol 6: 106-115.