Journal of Cytology & Molecular Biology
Download PDF
Date of receipt of sample Type of sample collected Patient’s ID Pathological report Results of the previous Immunohistochemical.
Research article
Quality of Sample in theMolecular Determination of Human Papillomavirus in Breast Tissue: Pre-Analytical and Analytical Error-Based Approach
Jeel Moya-Salazar*
- School of Medicine, Faculty of Health Sciences, Universidad Norbert Wiener, Peru
*Address for correspondence: School of Medicine, Faculty of Health Sciences, Universidad Norbert Wiener, Peru, Tel: +51 2010400;E-mail: jeel.moyasalazar@icloud.com
Citation: Moya-Salazar J. Quality of Sample in the Molecular Determination of Human Papillomavirus in Breast Tissue: Pre-Analytical and Analytical Error- Based Approach. J Cytol Molecul Biol. 2018;3(1): 6.
Journal of Cytology & Molecular Biology | ISSN: 2325-4653 | Volume: 3, Issue: 1
Submission: 10 October, 2018| Accepted: 12 November, 2018 | Published: 19 November, 2018
Copyright: © 2018 Moya-Salazar J. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Introduction: High-risk Human Papillomavirus (HR-HPV) can play an important role in the development of breast cancer. The investigations that attempt to clarify this purpose require quality in their results, which constitute the success of each study and the assurance of each result in both clinical practice and scientific research.
Objective: to evaluate the quality of breast tissue during the analysis of HR-HPV in samples of patients with breast cancer, in order to establish a quality control of pre-analytical and analytical processes in the laboratory of molecular biology useful for clinical diagnosis and for scientific research.
Materials and methods: Twenty-five samples of mammary carcinoma (12 surgical specimens and 13 biopsies) were included with prior informed consent at INEN in Peru, which had a diagnostic pathologic report with breast receptors (RE, RP and HER2) and were analyzed for HR-HPV with the My09/My11 system. The quality evaluation was performed during extraction and amplification (β-globin, positive control (HPV 16) and negative), and the sample quality analysis under CLSI guidelines MM13-A and CLSI MM06-A2.
Results: All samples were negative for HR-HPV and one had no β-globin amplification. An overall DNA concentration of ≤ 45 ng/μL was determined. Biopsies performed better than surgical specimens did (p=0.001). No association was found between samples with ≥ 1 positive breast marker with sample quality (p=0.588), or with molecular result (p=0.778).
Conclusion: Verification of breast tissue quality during the determination of HR-HPV in breast cancer showed poor quality, with low levels of DNA concentration and significant differences between sample types.
Keywords
Quality control; Human papilloma virus; Breast cancer; Cancer; Tissue preservation; Molecular biology
Introduction
Human papillomavirus (HPV) infection is the most common sexually transmitted infection worldwide [1]. The HPV infection is the necessary but not sufficient cause for the development of Cervical Cancer (CC), the main cause of female mortality and a public health problem in low-and-middle income countries [2,3]. There are more than 150 types of HPV, each with a particular tropism for specific anatomical sites, where High-Risk genotypes (HR-HPV), mainly genotype HPV 16 and HPV 18, cause ≥70% CC [4,5].
Recently several studies have cataloged that HPV-AR can play an important role in the development of breast cancer assuming a cosmopolitan distribution among communities [6,7]. In fact, this association has been identified in more than 17 countries in the five continents [6-11]. These findings and their future discoveries are of great scientific interest worldwide. The development of these investigations requires quality in their results, which constitute the success of each study and the assurance of each result both in clinical practice and in scientific research [12].
The molecular methods used for the identification of HPV in breast tissue and breast cancer were INNO-Lipa [9], the next Generation Sequencing, the Line Probe Assay reverse Hybridization system, In Situ Polymerase Chain Reaction (PCR), the Restriction Fragment Length Polymorphism [13-16], semi-nested PCR, and Real-Time PCR (RT-PCR) [6].
A prior all these evaluation methods depend on the pre-analytical phase. In this phase where the specimens are obtained or part of them, and they are stored, transported and processed analytically, the greatest number of technical errors have been reported [3,17]. Both the pre-analytical phase and the selection of the method of analysis are key factors in the detection of HPV due to final sensitivity and specificity of the methods [18].
In this sense, we aimed to verify the quality of breast tissue during the determination of the Human Papilloma Virus in breast cancer tissues. This objective was given in the framework of the establishment of a quality protocol for pre-analytical and analytical processes in the molecular biology laboratory useful for clinical diagnosis and for scientific research.
Materials and Methods
Samples
The selected samples were collected by biopsy and surgical specimens, with prior informed consent authorized by the ethics committee of the National Institute of Neoplastic Diseases (INEN) of Peru, as part of the objectives of the Cancer Research-Circle. The samples were collected in outpatient Oncology Care offices, which were referred to the Tumor Bank for storage in vials of 3ml at -60 °C in the freezing system until they were processed together. A standardized code for each sample was assigned.
The clinical records were collected to identify the main components of interest of the Cancer Research Circle:
Pathological report
Based on the clinical and pathological results, we have established the diagnosis of lobular, infiltrating, papillary breast carcinoma, etc. were established. We performed the evaluation of breast-hormonal recipients (Estrogen Receptor (ER), Progesterone Receptor (PR), and (human epidermal growth factor receptor 2 (C-Erb2 or HER2)) by Immunohistochemistry (Dako, Glostrup, Denmark) [19]. We included 25 samples of breast tissue for the evaluation of HPV infection and its immunological component.
Molecular test for HPV detection
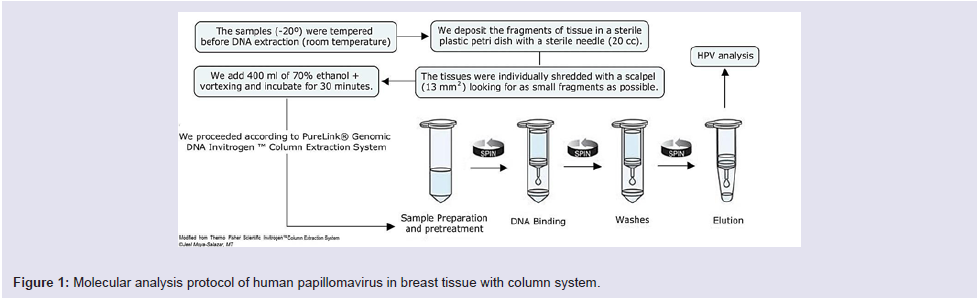
All the vials containing breast tissues were transported from the Tumor Bank to the area of Molecular Biology of the “Maes Heller” Center. We used the PureLink® Genomic DNA InvitrogenTM Column Extraction System (Thermo Fisher Scientific, Carlsbad, CA) extraction kit [20,21], as described in Figure 1.
The DNA extracts were quantified with the Qubit™ fluorometer (Thermo Fisher Scientific, Carlsbad, CA) with a cut-off value of 70 µl of DNA. The HPV’s DNA detection from breast tissue was performed with the HPV Consensus PCR and Genotyping system, which uses the My09/My11 primers that pair with the L1 region of the HPV genome (band 450 bp). In all analyzes (run of 5 samples) a pair of primers for the β-globin gene was included as an internal quality control.
The protocol indicated 2 cycles at 94 °C (30 sec), 3 cycles at 56 °C (35 sec), 4 cycles at 68 °C (35 sec), then 2 cycles at 94 °C (30 sec), 3 cycles at 56 °C (35 sec), and lastly 4 cycles at 68 °C (18 sec). Genotyping was performed with reverse line hybridization technique and chemiluminescence as described previously [21].
The amplified products were run by horizontal electrophoresis using agarose gel (3% at 112 V for 40 minutes) then colored with SYBR Safe (for 15 minutes) and visualized in the UV transilluminator.
Quality evaluation
In principle, we developed an amplification and extraction control process (primers for the human β-globin gene (GH20 (forward) - GAAGAGCCAAGGACAGGTAC, and corresponding PCO4 (reverse) -CAACTTCATCCACGTTC ACC) with a band of 268 bp). Moreover, a positive control (biopsy corresponding to HPV type 16 of 310 bp) and a blank as a negative control were included [21]. For the analysis of sample quality and storage, the evaluation parameters referred to in the CLSI guide MM13-A, MM06-A2, and the requirements for breast tissue from the American College of Pathologists were used [22-24].
Data analysis
The data analysis was performed with descriptive statistics, obtaining percentages, means and standard deviation values. We evaluated the associations between receptor negativity, type of sample, and type of breast carcinoma, and DNA quantification. In addition, we analyzed the variability between surgical specimen and biopsy related to the DNA quantification. The Pearson test was used considering a p value < 0.05 as statistically significant. The technique used for the statistical verification of the results was through the analyzer IBM SPSS v21.0 (Armonk, USA). This study received the approval of the INEN ethics committee as part of the objectives of the Cancer Research Circle (204-2015-FONDECYT).
Results
We analyzed 25 samples, of which 12 (48%) were surgical pieces, and 13 (52%) were biopsies with Core-Needle Aspiration (CNA). All these samples were extracted and analyzed in accordance with clinical requirements and pathological reports, all of which were negative for HPV infection.
The CNA had better performance than the surgical pieces (p = 0.001), obtaining 30% more nucleic acids Table 1. We was determined an overall concentration of DNA < 45 ng/ μL; DNA was extracted from 100% of samples.
Table 1: Major baseline components evaluated during the analysis of Human Papillomavirus in breast tissue of patients diagnosed with breast carcinoma.
The division includes the samples with histopathological diagnosis of invasive carcinoma of the breast, infiltrating carcinoma of breast subtype Nos / NST, infiltratingcarcinoma of the breast, infiltrating mammary carcinoma subtype NOS, breast tissue with the presence of focal atypical ductal hyperplasia, breast parenchyma with focal changes fibriadenomatous, chronic granulomatous mastitis, and fibroepithelial lesions related to fibroadenoma with the presence of ductal hyperplasia. ‡ Average concentration of DNA quantified with the fluorometer Qubit™ † Positive results: > 70% of cells with reactivity for the marker compared to the control. Abbreviations: ER: Estrogen Receptor; PR: Progesterone Receptor; HER2: Human Epidermal Growth Factor Receptor 2; n: Number of samples.
Table 1 shows the main quality components of the study. We showed fragments of different sizes that depended on the tissue collection method. Twenty-eight percent (7 samples) of CNA and surgical pieces had therapeutic margins. We found an association between tissues obtained with therapeutic margins and a low amount of DNA (rho=0.758, p=0.015).
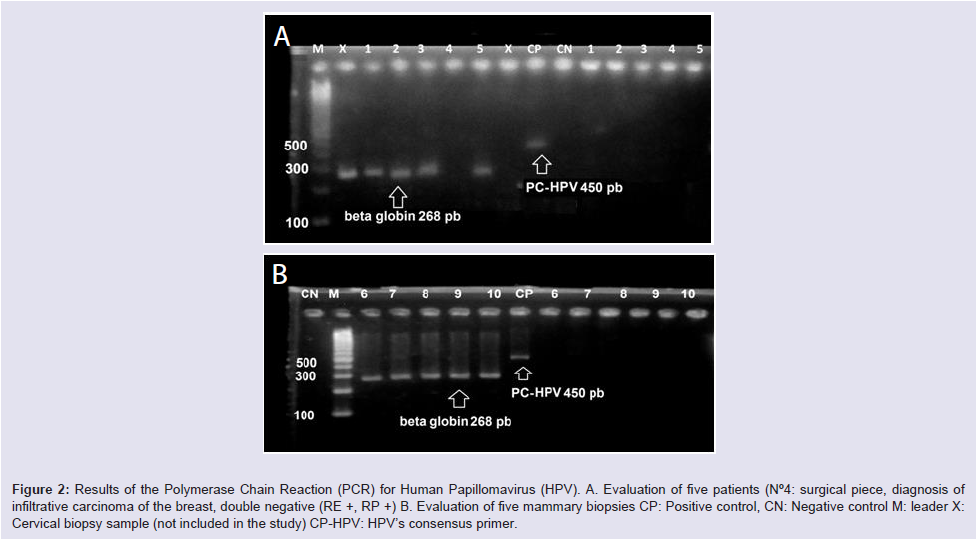
All positive controls had amplification at the level of the 450 band, with the exception of sample ID-4 that did not show amplification of the control gene (β-globin) being excluded Figure 2.
Figure 2: Results of the Polymerase Chain Reaction (PCR) for Human Papillomavirus (HPV). A. Evaluation of five patients (N°4: surgical piece, diagnosis of infiltrative carcinoma of the breast, double negative (RE +, RP +) B. Evaluation of five mammary biopsies CP: Positive control, CN: Negative control M: leader X: Cervical biopsy sample (not included in the study) CP-HPV: HPV’s consensus primer.
Four (16%) results were considered doubtful in their Immunohistochemical evaluation. In total, 3 triple-receptornegative samples were reported, 4 (16%) did not present pathological evaluations and 3 (12%) presented dubious results for theimmunohistochemical markers.
No association was found between the positivity of ≥1 breasthormonal- receptor marker with the sample quality (p=0.588), nor with the molecular result (p=0.778). No significant differences were found between the types of samples (p = 0.779) but in the clinicalpathological diagnosis (p = 0.017).
Discussion
In this study we showed that all tissue samples from breast cancer were negative for HPV. We determined that a sample did not demonstrate DNA extraction and amplification quality with the β-globin control, that 44% samples had low DNA levels (≤ 34 ng/μL) and that these characteristics were related to the type of sampling.
Molecular analysis in pathology is characterized by the dynamic change of technologies and markers, where the quality and quantity of nucleic acids are greatly affected by the type of collection, sample storage, manual processing and extraction method. There are real and very important differences between the analysis of HPV in cell brushes and breast tissue obtained by surgical procedures. In previous studies of cervical HPV-analysis, no great limitations have been found in the quantification acid nucleic [21], however, tissue and cellular evaluations depend on the number of altered cells present in the test sample, obtaining better results (reduction of false-negative results rates) with ≥ 200 cells per test sample [25].
The sampling is a critical process to ensure the integrity and accuracy of the quality of nucleic acids, since any inappropriate process results in the degradation of nucleic acids [22-24]. In the complexity of a high-performance pre-analytical phase, the validation and verification of the tests must be performed based on the requirement of a number of samples, which include the evaluation of the type and complexity of the test, the prevalence of the study’s goal in the community, data analysis requirements, etc. [26].
The type of study sample (smear, Formalin-Fixed Paraffin- Embedded (FFPE) tissue), anatomical location and population distribution affect the analysis of nucleic acids, such as HPV DNA [27,28]]. An inadequate DNA quality (degradation or fractionation) has been demonstrated in the analysis of HPV samples in tissues by PCR [29]. For breast tissue, the extraction methods are diverse (lupectomy, CNA, mastectomies, biopsies, surgical pieces with/ without margins, re-excision pieces, sentinel lymph node, etc.). All of these must ensure the correct pathological diagnosis, the estimation of prognostic factors, and response to treatment. The type of sample depends mainly on the previous clinical considerations and on the protocols that are followed before their diagnostic analysis; this represents a key point for the viral genomic analysis in breast tissue [30].
In the Peruvian clinical practice, the CNA and the surgical specimens with margins are the most frequent samples. For that reason, we need the importance of the correct fixation in neutral buffered formalin that avoids the false negatives and allows the visualization of the histological grade, the evaluation of vascular invasion, and other cellular components. Hence, we propose as an interference role the ink used to delimit the surgical margins, further investigation is also required to determine how the dye/ink impacts the analysis of subsequent nucleic acids.
Consideration should be given to the tissue fixation solution used during sampling since these usually induce DNA degradation [31]. Although the tissues can be stored for ≤ 2 years at ≤ 70 °C in liquid nitrogen, as was done in this study, not all health centers have these storage tools, so it is recommended to send the tissue immediately to both the pathological and research laboratory.
Due to the institution where the study was conducted presents strict work protocols, we could not perform pre-treatments to the breast tissues analyzed. These pre-treatments eliminate the inherently large amount of endogenous nucleases in tumor tissues, and the bloody components prior to analytical processing, as well as improve storage and processing time controls after thawing [22].
During the last decade, mainly the tissue biobanks have elaborated protocols for the maintenance of the integrity of the tissues and the preservation of the nucleic acids and proteins. The possibility of error results may be due to the low concentration of viral DNA in clinical samples, mainly in tissues or also, in the presence of endogenous PCR inhibitors in the sample [32]. In this study, we determined average viral DNA concentrations below the cut-off. This could explain the final result of the molecular analysis (Table 1). Other errors include the use of inadequate/inefficient nucleic acid extraction methods.
The type of nucleic acid extraction system will depend on the objectives of the molecular analysis, since an extraction for molecular sequencing, will not be the same as for the qualitative diagnosis of nucleic acids. In this study, we used a column extraction system that has high performance, although its limitations in the extraction of DNA from FFPE tissues (column capture method) have been demonstrated [21,33]. According to quality recommendations, each molecular tissue processing must perform repetitions of positive and negative controls per run [34].
The quality assurance strategies during the nucleic acid extraction and amplification of this study were the addition of the negative control (ultra-pure water) to ensure that there is no contamination during the analysis, and the amplification of β-globin in the same reaction for controlling adaptation of the sample. In each molecular evaluation, it is necessary to use reference genes (beta-actin, HLA, GAPDH, β-globin, HPRT1, among others) that generate validation data and stability of the analysis together for each experimental milieu. As all these have a certain degree of variation, the validation of the selected gene for each molecular process is necessary.
Besides for the purpose of eliminating errors in the quantification of nucleic acids and avoiding variations in the efficiency of RT-PCR derived in the MIQE guidelines, which currently allows the pertinent selection of endogenous controls for these analyze, we also consider that during conventional PCR this selection is vital and necessary [35]. There is software (geNorm, RefGenes, Genevestigator, etc.), spreadsheets (Bestkeeper, REST, etc.), among other tools that allow carrying out statistical evaluations of a panel of reference genes for the process of validation and standardization of the protocols with the samples together [36].
Although, the validation can be an embarrassing process, it will allow obtaining guarantor quality results that imply avoiding repetitions, because working with results from erroneously analyzed data (due to poor selection of housekeeping genes) can be more expensive in the long term. For example, conventional analysis of β-globin are a determining factor for the inclusion of participants in studies of HPV prevalence [37,38]. We propose that reference guides should be established (such as the MIQE guide) and the use of software for conventional PCR analysis (mainly Open Source).
In addition to the previous pre-analytical considerations for the detection of viral DNA, the selection and adequate use of analysis methods are required. The ability of the methods to amplify different sizes and types of DNA fragments of specific genotypes of HPV is one of the main limitations of molecular tests. For example, when HPV DNA detection was compared by PCR it was shown that the MY09/ MY11 method (15% detection) had less detection than the GP5+/ GP6+ method (L1 region amplification) [39].
Zhebe and Wilander demonstrated that both methods have a similar sensitivity in the detection of HPV in cervical biopsies, but point out that MY09/MY11 presents less performance (≤ 3%), like the results of Remmerbach et al., in oral mucosal preparations [40,41]. In part this could explain our negative results. However, to demonstrate the efficiency of the method, external quality control and verification of the protocols used is required [22,23].
The main goal of the study was not the comparative analysis of PCR techniques with My09/My11 primers against other methods (RT-PCR, in situ PCR, GP5+/GP6+method, etc.) for the detection of HPV in breast tissue, thus, we cannot explain the performance of the test. In general, if the detection method has limitations, analytical errors. Analytical errors (false results) of detection will be generated that will affect the quality of the study, and the clinical management of the patients with negative results. These limitations must be considered to ensure high-quality results that allow their correct interpretation.
Recently, the findings of HPV in breast tissue have been validated with the simultaneous analysis of ≥ 3 molecular methods [6,42-44]. This should have been considered in the current Peruvian report on aHPV in breast tissue to ensure the quality of its findings [45].
To evaluate all these quality components, there are international regulatory organizations that provide a basic system for the evaluation of clinical laboratories. However, molecular tests are not specified under the general requirements of the CLIA’88 guide where the quality ranges for each procedure are established.
The quality requirements establish a channel between the practical specific quality requirements of the main organisms (CLIA, FDA, CMS, etc.) with the accuracy in the diagnostic tests. But to establish these requirements are needed, among other things, acceptable types of samples for analysis. About breast tissue and HPV viral DNA standard guidelines are not yet established. Each laboratory should establish its own policies and procedures for the method under evaluation, in coordination with accredited agencies [46].
Finally, the null frequency of HPV reported in this study may also be due to the limited etiological distribution of the virus as the cause of this neoplasm, and the inclusion of patients without previous reports of cervical lesion or HR-HPV infection [6,42,43,47]. Although, several international groups have demonstrated the presence of HPV in different non-epithelial or mucosal organs [6-11,13-15,18,48-50], and their possible genetic role for the development of breast cancer [7,51], a rigorous causal-association has not yet been found that supports its clinical evaluation and explains its mechanism of breast infection [36,52].
More studies are required to evaluate the oncogenic role of HPV in breast tissue, explaining its possible non-sexual dissemination and the tropism of viral subtypes. Likewise, if a diligent association is established between HPV and breast carcinoma, its routine diagnostic application and its importance for health should be argued, because if HPV analysis for CC has not yet been seriously applied in Peru, we believe that its application to be far away for breast cancer.
Conclusion
Verification of breast tissue quality during the analysis of HPV in breast cancer showed poor quality, with low levels of DNA concentration and significant differences between sample types.
This same quality system should be evaluated in a greater number of samples and confronted with other DNA extraction systems, since there are technologies in development that allow improving the performance of the studies. All these elements are significant for the development of the current molecular pathology in both diagnosis and research.
We consider that with each quality assessment activity, the analysis procedures will be enhanced through various activities of continuous improvement, looking for quality to be assured in each phase based on the protocols organized to establish a quality management of preanalytical and analytical processes in the laboratory of molecular biology useful for clinical diagnosis and for scientific research.
References
- Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV (2002) The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 55: 244-265.
- zur Hausen H (2006) Infections causing human cancer. Weinheim: Wiley-VCH.
- Moya-Salazar J, Rojas-Zumaran V, Torres-Martínez R, Rosas-Vargas L (2016) Calidad de los extendidos cervicouterinos dentro de la coloración de Papanicolaou para el cribado de cáncer cervical en Lima, Perú. Rev Esp Patol 49: 7-18.
- de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, et al. (2010) Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective crosssectional worldwide study. Lancet Oncol. 11:1048-56.
- Bosch FX, Broker TR, Forman D, Moscicki AB, Gillison ML, et al. (2013) Comprehensive control of HPV infections and related diseases. Vaccine 31: H1-31.
- Lawson JS, Glenn WK, Salyakina D, Clay R, Delprado W, et al. (2016) Human Papilloma Virus identification in breast cancer patients with previous cervical neoplasia. Front Oncol 5: 298.
- Ohba K, Ichiyama K, Yajima M, Gemma N, Nikaido M, et al. (2014) In vivo and in vitro studies suggest a possible involvement of HPV infection in the early stage of breast carcinogenesis via APOBEC3B induction. PLoS One 9: e97787.
- Simoes PW, Medeiros LR, SimoesPires PD, Edelweiss MI, Rosa DD, et al. (2012) Prevalence of human papillomavirus in breast cancer: a systematic review. Int J Gynecol Cancer 22: 343-7.
- Fernandes A, Pesci-Feltri A, García FA, Guida V, Salazar JM, et al. (2015) Evaluation of human papilloma virus infection in patients with breast cancer. Rev Venez Oncol 27: 22-29.
- Herrera-Goepfert R, Vela-Chávez T, Carrillo-García A, Lizano-Soberón M, Amador-Molina A, et al. (2013) High-risk human papillomavirus (HPV) DNA sequences in metaplastic breast carcinomas o Mexican women. BMC Cancer. 13: 445.
- Da Silva JRG (2010) Avaliação da prevalência do Papilomavírus Humano em carcinoma mamário. [Tesis] Teresina: Universidade Federal Do Piauí.
- Clinical and Laboratory Standards Institute (CLSI) (2013) Design molecular proficiency testing/External quality assessment; Approved guideline-Second edition. CLSI document CLSI MM14-A2. Clinical and Laboratory Standards Institute (CLSI). 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA.
- Li N, Bi X, Zhang Y, Zhao P, Zheng T, et al (2011) Human papillomavirus infection and sporadic breast carcinoma risk: a meta-analysis. Breast Cancer Res Treat 126: 515-520.
- Duo D, Ghimenti C, Migliora P, Pavanelli MC, Mastracci L, et al. (2008) Identification and characterization of human papillomavirus DNA sequences in Italian breast cancer patients by PCR and line probe assay reverse hybridization. Mol Med Rep 1: 673-677.
- Heng B, Glenn WK, Ye Y, Tran B, Delprado W, et al. (2009) Human papilloma virus is associated with breast cancer. Br J Cancer 101: 1345-50.
- Mahmoodi P, Motamedi H, Abad MRSS, Bahrami MS, Kargar M (2016) molecular detection and typing of human papillomaviruses in paraffin-embedded cervical cancer and pre-cancer tissue specimens. Iran J Cancer Prev 9: e3752.
- Plebani M (2012) Quality Indicators to Detect Pre-Analytical Errors in Laboratory Testing. Clin Biochem Rev 33: 85-88.
- Amarante MK, Watanabe MA (2009) The possible involvement of virus in breast cancer. J Cancer Res Clin Oncol 135: 329-337.
- Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, et al. (2010) American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med 134: e48-72.
- Concha-M R, Arias-Stella JJ, Quiñones D, Bazan M, Iwasaki R, et al. (2012) Investigacion del ADN del virus del papiloma humano en el cuello uterino en poblacion rural del Peru. Patol Rev Latinoam 50: 266-271..
- Sullcahuaman-Allende Y, Castro-Mujica M, Mejia Farro R, Castañeda CA, Castillo M, et al. (2015) Características sociodemográficas de mujeres peruanas con virus papiloma humano detectado por PCR-RFLP. Rev Peru Med ExpSalud Publica 32: 509-514.
- Clinical and Laboratory Standards Institute (CLSI) (2005) MM13-A- Collection, Transport, Preparation, and Storage of Specimens for Molecular Methods; Approved Guideline. CLSI document MM13-A. Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898 USA.
- Clinical and Laboratory Standards Institute (CLSI) (2010) Quantitative molecular methods for infectious diseases; approved guideline, 2nd Edition. CLSI document MM06-A2. Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898 USA.
- College of American Pathologist (2012) Molecular Pathology Checklist. Northfield: CAP Accreditation Program.
- Eltoum I, Chhieng DC, Crowe DR, Roberson J, Jin G, et al (2007) Significance and possible causes of False-negative results of reflex Human Parpillomavirus infection testing. Cancer 111: 154-59.
- Burd EM (2010) Validation of laboratory-developed molecular assays for infectious diseases. Clin Micro Rev 23: 550-576.
- Demathe A, Bernabé GD, Garcia JF, Nunes MC, Miyahara IG. (2010) Comparação entre dois métodos de detecção de DNA de papilomavírus humano em carcinoma epidermoide de lábio. J Bras Patol Med Lab 46: 85-90.
- Mee BC, Carroll P, Donatello S, Connolly E, Griffin M, et al. (2011) Maintaining breast cancer specimen integrity and individual or simultaneous extraction of quality DNA, RNA, and Proteins from Allprotect-Stabilized and nonstabilized tissue samples. Biopreser Biobanking 9: 389-398.
- Steiner HJ, Periger P, Moser P, Schmid T, Kern MA, et al. (2009) Improved ultrastructural resolution by a new re-embedding procedure for formaldehyde-fixed paraffin-embedded tissues. Histopathology 54: 775-777.
- SociedadEspañola de Ginecología y Obstetricia (SEGO), SociedadEspañola de AnatomíaPatológica (SEAP) (1999) Protocolos de estudio e informeanatomopatológico de tumoresmalignosginecológicos y mamários. Madrid: Nova Sidonia Oncología; 1999.
- Diaz-Cano SJ, Brady SP (1997) DNA extraction from formalin-fixed, paraffin-embedded tissues: Protein digestion as a limiting step for retrieval of high-quality DNA. Diagn Mol Pathol 6: 342-346.
- Vince A, Poljak M, Seme K (1998) DNA extraction from archival giemsa-stained bonemarrow slides: comparison of six rapid methods. Br J Haematol 10: 349-51.
- Huijsmans CJJ, Damen J, van der Linden JC, Savelkoul PHM, Hermans MHA (2010) Comparative analysis of four methods to extract DNA from paraffin-embedded tissues: effect on downstream molecular applications. BMC Res Notes 3: 239.
- Jennings L, Van Deerlin VM, Gulley ML (2009) Recommended principles and practices for validating clinical molecular pathology tests. Arch Pathol Lab Med. 133: 743-755.
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611-622.
- Vandesompele J, De Preter K, Pattyn F, , Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034.
- Del Rio-Ospina L, Soto-De Leon SC, Camargo M, Sanchez R, Mancilla CL, et al. (2016) The Prevalence of High-Risk HPV Types and Factors Determining Infection in Female Colombian Adolescents. PLoS One 11: e0166502.
- Murillo R, Molano M, Martinez G, Mejia JC, Gamboa O (2009) HPV prevalence in Colombian women with cervical cancer: implications for vaccination in a developing country. Infect Dis Obstet Gynecol: 653598.
- Meneses VE, Muniz BM, Mota LA, Souza VA, Casimiro OAS, et al. (2014) HPV detection using primers MY09/MY11 and GP5+/GP6+ in patients with cytologic and/or colposcopic changes. J Bras Patol Med Lab 50: 280-285.
- Zehbe I, Wilander E (1996) Two consensus primer systems and nested polymerase chain reaction for human papillomavirus detection in cervical biopsies: a study of sensitivity. Hum Pathol 27: 812-815.
- Remmerbach TW, Brinckmann UG, Hemprich A, Chekol M, Kühndel K, et al (2004) PCR detection of human papillomavirus of the mucosa: comparison between MY09/MY11 and GP5+/6+ primer sets. J ClinVirol 30: 302-308.
- Hennig EM, Suo Z, Thoresen S, Holm R, Kvinnsland S, et al. (1999) Human papillomavirus 16 in breast cancer of women treated for high-grade cervical intraepithelial neoplasia (CIN III). Breast Cancer Res Treat. 53: 121-135.
- Widschwendter A, Brunhuber T, Wiedemair A, Mueller-Holzner E, Marth C. (2004) Detection of human papillomavirus DNA in breast cancer of patients with cervical cancer history. J ClinVirol 31: 292-297.
- Salman NA, Davies G, Majidy F, Shakir F, Akinrinade H,et al. (2017) Association of High Risk HumanPapillomavirus and Breast cancer:A UK based Study. Sci Rep 7: 43591.
- Galvez M, Belmar-Lopez C, Calderon G, Sanchez J, Castillo GM, (2018) Presence of human papillomavirus in breast cancer and its association with clinical features. J Clin Oncol 36: e13566.
- Clinical and Laboratory Standards Institute (CLSI) (2013) Measurement Procedure Comparison of Bias and Estimation Using Patients Samples; Approved Guideline-Third Edition. CLSI document CLSI EP9-A3. Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898 USA.
- Hedau S, Kumar U, Hussain S, Shukla S, Pande S,et al (2011) Breast cancer and human papillomavirus infection: No evidence of HPV etiology of breast cancer in Indian women. BMC Cancer 11: 27.
- Whitaker NJ, Glenn WK, Sahrudin A, Orde MM, Delprado W, et al. (2012) Human Papillomavirus and Epstein Barr Virus in Prostate Cancer: Koilocytes Indicate Potential Oncogenic Influences of Human Papillomavirus in Prostate Cancer. Prostate 73: 236-241.
- Mucino HMI (2010) Identificacion del Virus de Papiloma Humano en neoplasia de tiroides. [Thesis de Maestria]. México: Facultad de Medicina, Universidad de Colina.
- Termine N, Panzarela V, Falaschini S, Russo A, Matranga D, et al. (2008) HPV in oral squamouscell carcinoma vs head and necksquamouscells carcinoma biopsies: a meta-analysis (1988-2007). Ann Oncol 19: 1681-690.
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, et al. (2013) APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 21: 366-370.
- Bodaghi S, Wood LV, Robby G, Ryder C, Steiberg SM, et al. (2005) Could Human Papillomavirus be spread through blood? J ClinMicrobiol 43: 5228-5234.